-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA New Glycan-Dependent CD4-Binding Site Neutralizing Antibody Exerts Pressure on HIV-1
CD4bs is a central viral vulnerability site and isolation of new anti-HIV-1 CD4bs broadly neutralizing antibodies (bNAbs) provides information about viral escape mechanisms. Here we describe a new anti-HIV-1 bNAb that was isolated from an HIV-1 infected donor. The antibody, 179NC75, targets the CD4 binding site in a glycan-dependent manner. Although many CD4bs antibodies have been already described, a glycan-dependent mode of recognition is unusual for anti-HIV-1 CD4bs bNAbs. The glycan-dependent CD4bs antibodies have never been tested for their ability to neutralize HIV-1 in vivo. We infected humanized mice with HIV-1YU2 and treated them with 179NC75 three weeks after infection. We observed a drop in viral load immediately after treatment followed by a viral rebound. The viral rebound was associated with specific escape mutations in the plasma virus envelope, resulting in a deletion of N276 glycan, and in some cases a glycan shift from position 276 to position 460. Similar signature mutations were found in the envelope of the autologous virus cloned from patient’s plasma. This defines the escape pathways from 179NC75, and shows that they are the same in humans and in HIV-1YU2 infected humanized mice.
Published in the journal: . PLoS Pathog 11(10): e32767. doi:10.1371/journal.ppat.1005238
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005238Summary
CD4bs is a central viral vulnerability site and isolation of new anti-HIV-1 CD4bs broadly neutralizing antibodies (bNAbs) provides information about viral escape mechanisms. Here we describe a new anti-HIV-1 bNAb that was isolated from an HIV-1 infected donor. The antibody, 179NC75, targets the CD4 binding site in a glycan-dependent manner. Although many CD4bs antibodies have been already described, a glycan-dependent mode of recognition is unusual for anti-HIV-1 CD4bs bNAbs. The glycan-dependent CD4bs antibodies have never been tested for their ability to neutralize HIV-1 in vivo. We infected humanized mice with HIV-1YU2 and treated them with 179NC75 three weeks after infection. We observed a drop in viral load immediately after treatment followed by a viral rebound. The viral rebound was associated with specific escape mutations in the plasma virus envelope, resulting in a deletion of N276 glycan, and in some cases a glycan shift from position 276 to position 460. Similar signature mutations were found in the envelope of the autologous virus cloned from patient’s plasma. This defines the escape pathways from 179NC75, and shows that they are the same in humans and in HIV-1YU2 infected humanized mice.
Introduction
Although the envelope glycoproteins (Env) of primate immunodeficiency viruses have extremely variable sequences [1], most of them engage CD4 as the primary cellular receptor to initiate the viral life cycle [2]. The consequence is that the CD4 binding site (CD4bs) is a comparatively well-conserved region of Env that serves as a critical neutralization epitope and an appealing vaccine target. The introduction of single cell antibody cloning techniques [3,4] yielded dozens of broad and potent CD4bs antibodies from infected individuals, some of which neutralize ~90% of HIV-1 strains in vitro [5–7]. Some of these antibodies are also effective at reducing viral load when used to treat infected humanized mice (hu-mice) [8], macaques [9–11] and humans [12].
The most potent group of CD4bs antibodies characterized to date is derived from two VH genes, IGVH1-2 [5,7,13] and IGVH1-46 [6,7,14–16]. These antibodies engage many of the same Env residues as CD4. For example, residue Arg71HC in VRC01-like bNAbs interacts with residue Asp368gp120 on Env, and thereby mimics how Arg59CD4 interacts with the same residue when CD4 binds to gp120 [6,7,13,16]. Although the light chains are less restricted in their origin, specific alterations are required for activity, including mutations and deletions [6,13,16]. Overall, the restricted origins and complex development of these bNAbs from their inactive germline precursors may explain why it has been so difficult to elicit them by vaccination.
A second, far more diverse group of CD4bs-directed antibodies is often referred to as ‘CDRH3-dominated class of CD4bs antibodies’. These antibodies use their CDRH3-loop regions to engage Env [15]. These include b12 [17], HJ16 [18], CH103 [19] and the recently described VRC13 and VRC16 [15]. Structural analyses indicate that all CDRH3-dominated antibodies use loop-based recognition mechanisms, with the CDRH3 contributing 50%-70% of the paratope interface [15,19,20]. They are not VH-restricted since their CDRH3s are randomly assembled from IgH variable, diversity and joining segments during V(D)J recombination [21]. In keeping with their diverse origins, CDRH3-dominated antibodies seem to employ different mechanisms of recognition and they also vary in the angles with which they approach the CD4bs [15].
To isolate new CD4bs bNAbs, we sought HIV-1 infected donors whose sera contained potent neutralizing antibodies that appeared to target the CD4bs. One such donor was EB179. By sorting peripheral blood mononuclear cells (PBMCs) from this individual we isolated a new antibody, 179NC75, that is encoded by IGVH3-21 and IGVL3-1 gene segments. In TZM.bl neutralization assays 179NC75 showed an overall IC80 of 0.42 μg/ml against 120 Tier-2 HIV-1.
Binding assays using various Env-based proteins indicated that 179NC75 is glycan-dependent and belongs to the same sub-class of CDRH3-dominated CD4bs antibodies as HJ16. These glycan-dependent CD4bs antibodies have not yet been tested for activity in vivo. To do so we treated humanized mice infected with HIV-1YU2 with 179NC75 and found that it selects for escape variants with mutations in the potential N-linked glycosylation site at gp120 position 276. Similar mutations were also found in the autologous isolate from the 179NC75 donor, suggesting that selection pressure had been exerted in the human host.
Materials and Methods
Ethics statement
For the human studies, The Rockefeller University Institutional Review Board approved all studies involving patient enrollment, sample collection, and clinical follow-up. Donor EB179 was selected from a group of long-term non - progressors that was followed at the Ragon Institute in Boston, and is also referred to as subject 330183. The subject described in this study provided written informed consent prior to participating in this study. For the mouse studies, this study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of The Rockefeller University, and in accordance with established guidelines and policies at The Rockefeller University (protocol number 13618-H).
HIV-1-infected subjects
Purified IgG samples from 394 HIV-1-infected long-term non-progressors were screened for neutralizing activity against a panel of 14 viruses representing 8 different clades or inter-clade recombinants. IgG purified from donor EB179 was exceptional in its neutralization potency and breadth, ranking within the top 2% of the cohort, and neutralized 11 out of the 14 viruses in the panel (S1A Table). A single leukapheresis sample was obtained 4.5 years after initial diagnosis with clade B HIV-1 infection, at age 44. At the time the sample was collected, donor EB179 had 1038 CD4 T cells/mm3 and a viral load of 3180 copies/ml and was not receiving antiretroviral therapy. Molecular HLA typing revealed HLA A*02 : 01, 68 : 02; B*07 : 02, 53 : 01; Cw04 : 01, 07 : 02; DRB11 : 01, 15 : 01.
Single B cell sorting and antibody cloning
Single-cell sorting of 2CC core+CD19+IgG+ B cells from donor EB179’s PBMCs was conducted as previously described [3]. Briefly, we sorted memory B cells using the gp120 2CC core protein as bait [22]. Rescue primers were used to amplify both heavy chains [7] and Igλ genes [23]. All PCR products were sequenced and analyzed for Ig gene usage, CDR3, and the number of VH/VL somatic hypermutations (IgBLAST, http://www.ncbi.nlm.nih.gov/igblast/ and IMGT, http://www.imgt.org/). Multiple sequence alignments were performed using the MacVector program (v.13.5.5) with the ClustalW analysis function (default parameters), and then used to generate dendrograms by the neighbor-joining method (with best tree mode and outgroup rooting). To specifically isolate members of the 179NC75 clone we used the following forward primers for the heavy chains: 5’-CTGCAACCGGTGTACATTCTGAAATGAGATTGGAAGAAT-3’ and 5’-CTGCAACCGGTGTACATTCTGAGGTCCAGTGTGAAGAA-3’ (in a 1 : 1 mix); and for the light chains: 5’-ATGGCCTGGATCCCTCTACTTCTC-3’ and 5’ - ATGGCATGGATCCCTCTCTTCCTC-3’ (in a 1 : 1 mix). The reverse primers were the same as described previously for Ig gene amplification [7].
Antibody production
Purified, digested PCR products were cloned into human Igγ1-, IgK or Igλ-expression vectors as previously described [24]. Antibodies were produced by transient transfection of IgH, IgK and IgL expression plasmids into exponentially growing HEK 293T cells (ATCC; CRL-11268) using polyethyleneimine (PEI)-precipitation [24]. IgG antibodies were affinity purified using Protein G Sepharose beads according to the manufacturer’s instructions (GE Healthcare).
ELISAs
High-binding 96-well ELISA plates (Costar) were coated overnight with 5 μg/ml of purified 2CC core, gp120.YU2 (wild type or mutants) or gp140.YU2 foldon trimer in PBS. After washing 6 times with PBS + 0.05% Tween 20, the plates were blocked for 2 h with 2% BSA, 1 μM EDTA and 0.05% Tween-PBS (“blocking buffer”), and then incubated for 1 h with IgGs that were added as seven consecutive 1 : 4 dilutions in PBS from an initial concentration of 4 μg/ml. After additional washing, the plates were developed by incubation with goat HRP-conjugated anti-human IgG antibodies (Jackson ImmunoResearch) (at 0.8 μg/ml in blocking buffer) for 1 h followed by HRP chromogenic substrate (ABTS solution; Invitrogen). For competition ELISAs, the plates were coated with 5 μg/ml 2CC core, gp120 or gp140 foldon, washed, blocked for 2 h with blocking buffer and then incubated for 1 h with IgGs added as seven consecutive 1 : 4 dilutions in PBS from an initial concentration of 32 μg/ml, and in the presence of biotinylated 179NC75 antibody at a constant concentration of 4 μg/ml. The plates were then developed using HRP-congugated streptavidin (Jackson ImmunoReseach) (at 1 μg/ml in blocking buffer).
For ELISAs using BG505 SOSIP.664-D7324 trimers, the plates were coated overnight with 5 μg/ml of D7324 antibody as previously described [25], washed and then incubated with 500 ng/ml of the trimer [25,26]. After a further wash, IgGs were added for 1 h as seven consecutive 1 : 4 dilutions in PBS from initial concentrations of 4 μg/ml. The endpoint was generated by incubation with goat HRP-conjugated anti-human IgG antibodies, as described above. All experiments were performed at least 3 times.
For EndoH ELISA, the plates were coated overnight at 4°C with 5 μg/ml of EndoH-treated or untreated gp120 in 100 mM sodium bicarbonate/carbonate buffer, pH 9.6. They were then washed with TBS + 0.05% Tween 20 and blocked for 1 h in the same buffer supplemented with 3% (w/v) BSA, and washed again before test antibodies were added for 2 h. After a final wash, the endpoint was generated using goat HRP-conjugated anti-human IgG antibodies, again as described above.
Neutralization assay
HIV-1 neutralization was evaluated using the luciferase-based TZM.bl cell assay as described previously [27]. Briefly, envelope pseudoviruses were incubated with fivefold serial dilutions of single antibodies and applied to TZM.bl cells that carry a luciferase-reporter gene. After 48 h cells were lysed and luminescence was measured. IC50 and IC80 reflect single antibody concentrations that caused a reduction in relative luminescence units (RLU) by 50% and 80%, respectively.
Humanized mice (hu-mice)
NOD Rag1−/−Il2rgnull (NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl/SzJ) mice were purchased from The Jackson Laboratory and bred and maintained at the Comparative Bioscience Center of The Rockefeller University according to guidelines established by the University’s Institutional Animal Care and Use Committee. All experiments were performed under protocols approved by the same committee. Hu-mice were treated with 1 mg of 179NC75 sub-cutaneously (s.c.) on day 0, followed by 0.5 mg s.c. injections twice-weekly for a period of 5 weeks [8]. The gp120 sequences from escape variant viruses were obtained as previously described [8].
Virus cloning and virus co-culture
The autologous virus from donor EB179 was isolated as previously described [28]. Briefly, CD19 and CD8-depleted mononuclear cells were cultured at a concentration of 5 × 106 cells/ml in Iscove's modified Dulbecco’s medium (IMDM; Gibco) supplemented with 10% fetal bovine serum (FBS; HyClone, Thermo Scientific), 1% GlutaMAX (Gibco), 1% penicillin/streptomycin (Gibco), and 1 μg/ml phytohaemagglutinin (Life Technologies) at 37°C and in an atmosphere containing 5% CO2. After 2–3 days, 5 × 106 cells were transferred into IMDM supplemented with 10% FBS, 1% penicillin/streptomycin, 5 μg/ml polybrene (Sigma), and 100 IU/ml of IL-2. The medium was replaced weekly and the HIV-1 content of culture supernatants was quantified using the Lenti-X p24 Rapid Titer Kit (Clontech) according to the manufacturer’s instructions. Env genes from the autologous virus were cloned by reverse transcriptase PCR as described elsewhere [29].
HIV-1YU2 envelope mutants
Single, double and triple mutations were introduced into wild-type HIV-1YU2 envelope using the QuikChange (multi-) site-directed mutagenesis kit, according to the manufacturer’s specifications (Agilent Technologies).
Results
Serologic specificity
Polyclonal IgG purified from donor EB179 had exceptional neutralization capacity, with respect of potency and activity against 11 of 14 Tier-2 viruses in a small cross-clade panel (S1A Table). To map the predominant NAb specificities, we tested EB179 IgG against HIV-1YU2 mutants that are resistant to NAbs targeting the trimer apex (N160K), the CD4bs (N280Y) or the base of the V3 loop (N332K) [8,30–32]. Among these mutants, only HIVYU2N280Y was resistant to EB179 IgG (S1B Table). We conclude that at least a proportion of the neutralization activity present in this serum is directed to the CD4bs.
EB179 CD4bs antibody repertoire
To isolate and characterize the NAbs present in EB179, we used flow cytometry to sort memory B cells that bound to 2CC core, a gp120 antigen that presents the CD4bs in an exposed and stable conformation [22]. Among CD19+IgG+ B cells, ~0.2% bound strongly to 2CC core. Of the 372 cells sorted, 87 produced paired heavy and light chains, 36 of which represented ten clonally related families (Fig 1A). Antibody sequences obtained from the expanded B cell clones contained higher numbers of somatic mutations compared to antibodies obtained from B cells that appeared only once (S1 Fig). The average number of nucleotide mutations in the heavy chain of clonal sequences was 44.76 (± 3.66, N = 36) compared to 20.82 (± 1.39 N = 51) for unique sequences (S1A Fig). A similar trend was observed when the light chain sequences were analyzed (S1B Fig).
Fig. 1. 2CC core antibody repertoire in patient EB179. 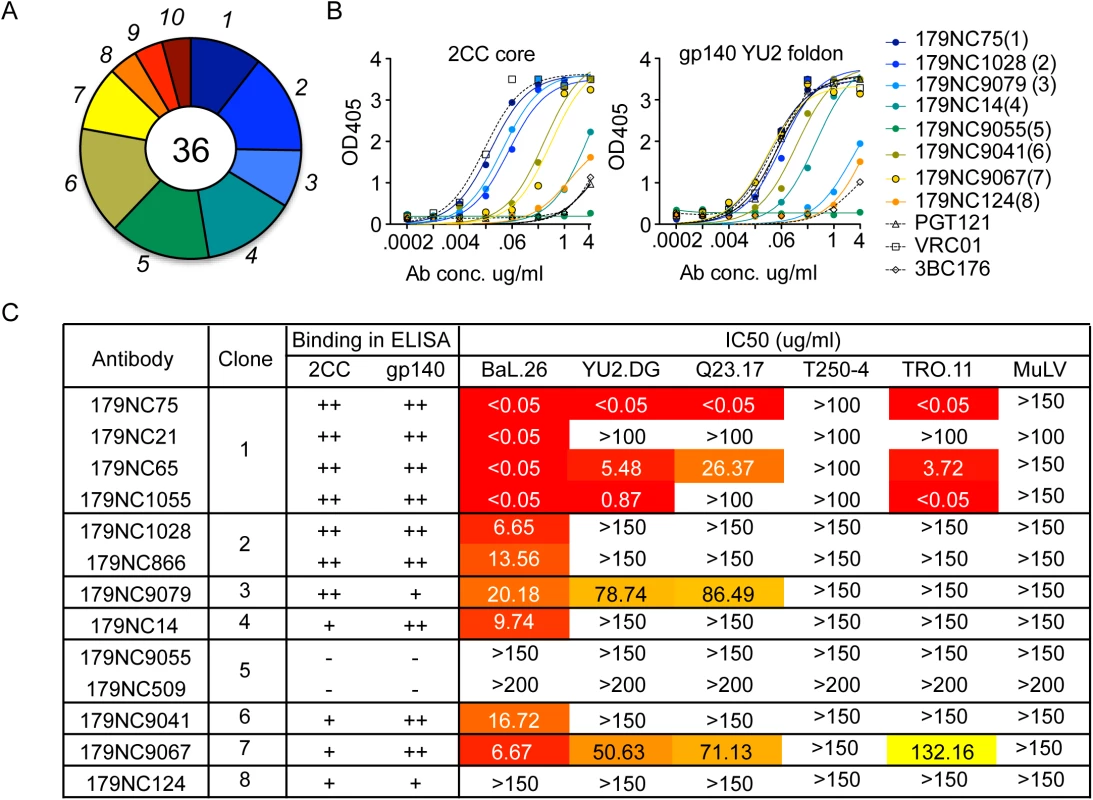
(A) Pie chart representing ten antibody sequence families sorted using the 2CC core protein as bait. The number in the center of the pie denotes the number of antibodies; slices are proportional to clone size (i.e., frequency) and represent unique clones. The assigned number of each clone is given next to the corresponding slice on the pie chart. The membership of an antibody in a B cell clone is determined by sequence analysis, in particular of the CDR3s and shared V and J genes of paired heavy and light chain genes (see Methods). (B) Representative variants of each clonal family of sequences were expressed and tested in ELISA for binding to 2CC core and gp140 foldon proteins. The numbers in parentheses correspond to the number of the clone as shown in (A). (C) The table summarizes the different antibodies expressed, the clone they represent and their binding to 2CC core or gp140 YU2 foldon (based on (B)) and their in vitro neutralization IC50 values, as measured in the TZM.bl cell assay. Representative variants from each of the clonal families were selected for further analysis (S2 Table). These variants were expressed as IgG1 antibodies that were tested for binding to a HIV-1YU2 gp140 foldon protein [33] or 2CC core [22], and for neutralizing activity. Except for 179NC9055, all the antibodies bound strongly to the HIV-1YU2 gp140 and/or 2CC core proteins (Fig 1B), and members of clones 1, 2, 3, 4, 6, and 7 neutralized the Tier-1 (i.e., neutralization-sensitive) HIV-1BAL virus (Fig 1C). While antibodies from clones 3 and 7 were only weakly active against the other viruses in the panel, one representative of the most expanded clone 1 (179NC75) strongly neutralized four of the five viruses tested (IC50 ≤0.05 μg/ml, Fig 1C).
179NC75 and its clonal relatives
To isolate additional 179NC75 variants, we amplified cDNA from the 2CC core+CD19+IgG+ single-sorted B cells using specific VH and VL forward primers (see Methods). We obtained a total of 23 heavy chain and 25 light chain variants from the 179NC75 clonal family. The heavy and light chain sequences carried 34% and 29% amino acid mutations on average, respectively, compared to their germline gene segments IGVH3-21 and IGVL3-1. The various sequences of the 179NC75 clone were similar by up to 73% from clonal members (Fig 2A and 2B). The CDRH3 and CDRL3 regions were 24 and 10 residues long, respectively (Fig 2A and 2B, S2 Table). There were no insertions or deletions.
Fig. 2. 179NC75 clonal family variants. 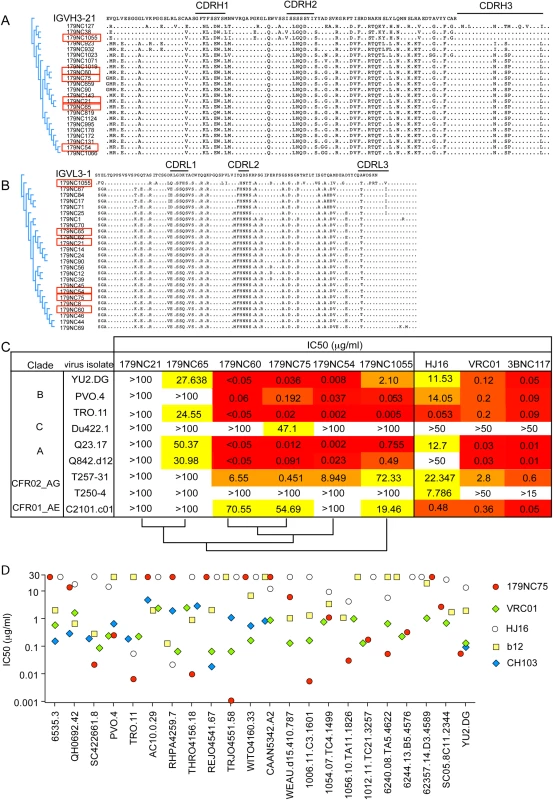
(A) Dendrogram showing heavy chain amino acid sequence alignment of the different 179NC75 variants to their closest germline IGVH3-21. (B) The same plot as in (A), but for the light chains. The heavy and light chains of the variants that were expressed and tested for neutralization are marked with red squares. (C) Neutralization IC50 values for 179NC75 and five variants against an expanded panel of Tier-2 viruses. The phylogenetic relationship, based on heavy chain sequences, between the six variants are shown on the bottom of the table (D) The plot compares the IC50 values for 179NC75 (red circles) and four well characterized CD4bs bNAbs (VRC01 –green diamonds, b12 –yellow squares, CH103 –blue diamonds and HJ16 –white circles) against a panel of 26 clade B Tier-2 viruses. Variants 179NC 54, 60, 65, 75, 21 and 1055 (indicated in Fig 2A and 2B) were tested for activity against a panel of nine Tier-2 viruses, including three from clade B, one from clade C, two from clade A, two clade A/G recombinants and one clade A/E recombinant. 179NC75 and two closely related variants, 179NC54 and 179NC60, potently neutralized 6 of these 9 viruses, whereas the other antibodies had lesser or no neutralization activity (Fig 2C). Accordingly, we selected 179NC75 for additional analyses.
When tested against an extended cross-clade panel of 120 Tier-2 viruses, 179NC75 neutralized viruses from clades B particularly strongly (S3 Table); its geometric mean IC50 and IC80 values were 0.113 μg/ml and 0.291 μg/ml, respectively (S4 Table). When compared to other CD4bs bNAbs against a panel of 22 Tier-2 clade, B viruses, 179NC75 was more potent than b12 against 13 viruses, than HJ16 against 15 viruses, than VRC01 against 8 viruses, and than CH103 against 6 viruses (Fig 2D). Its overall breadth of activity across the clade B virus panel was 70% (S4 Table).
179NC75 binds Env by an HJ16-type mechanism
To map the epitope targeted by 179NC75 and its clonal variants, we performed a series of ELISAs. All members of the 179NC75 clonal family bound to HIV-1YU2 gp120, gp140 foldon [34] and 2CC core [22] proteins (S2 Fig). In a competition ELISA, soluble CD4 (sCD4) and most CD4bs antibodies competed with 179NC75 for binding to gp120YU2, whereas PGT121, PGT128 and 10–1074 did not (Fig 3A upper and lower panels). The 8ANC195 bNAb, which binds an epitope adjacent to the CD4bs [7,35], inhibited 179NC75 binding by ~ 50% (Fig 3A, lower panel). We conclude that the 179NC75 epitope is proximal to the CD4bs.
Fig. 3. Epitope mapping of 179NC75 variants by ELISA. 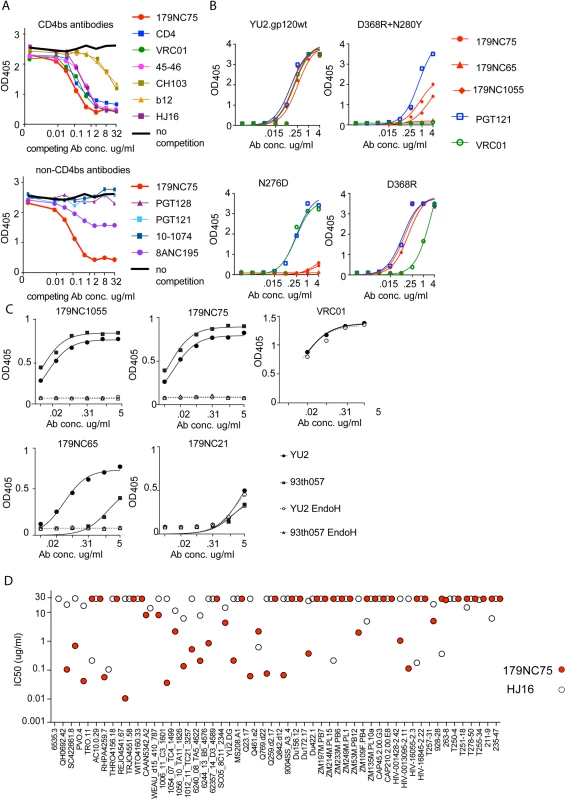
(A) Competition ELISA. The plots show the binding of biotinylated 179NC75 (constant concentration of 2 μg/ml) to gp120 in the presence of increasing concentrations of different bNAbs. The black bold line represents 179NC75 binding in the absence of a competitor antibody, the red bold line shows auto-competition by non-biotinylated 179NC75. Upper panel: Competition by CD4bs bNAbs; lower panel competition by non-CD4bs bNAbs. (B) Binding of 179NC75 (red circles), 179NC65 (red triangles) and 179NC1055 (red diamonds) antibodies to the wild-type YU2 gp120 monomer (left upper panel) and variants containing CD4bs-related mutations (D368R+N280Y, right upper panel; D368R, right lower panel; N276D, left lower panel), VRC01 and PGT121 served as control antibodies. (C) 179NC75, 179NC65 and 179NC1055 binding to gp120 monomers from YU2 (clade B) and 93TH057 (clade A/E) before and after deglycosylation with EndoH. VRC01 was used as a control antibody in the experiment involving the YU2 gp120s. (D) Comparison between the neutralizing activity (IC50) of 179NC75 and HJ16 against a cross-clade virus panel of Tier-2 viruses. We next tested how different mutations in the CD4bs affected 179NC75 binding. The D368R single mutation was not sufficient to affect the gp120-binding of 179NC75 family members, but the D368R and N280Y double mutation substantially impaired their binding. In contrast, VRC01 is sensitive to the single D368R substitution (Fig 3B, right upper and lower panels).
The Asn276 glycan site is important for the binding of two different bNAbs: the CD4bs antibody HJ16 [36] and the gp120-gp41 specific antibody 8ANC195 [7,35]. The 8ANC195 epitope lies outside the CD4bs and this antibody binds Env in the presence of CD4 [7,35]. Since HJ16 strongly inhibited 179NC75 binding (Fig 3A, upper panel) and 8ANC195 did so weakly (Fig 3A, lower panel), we assessed whether the binding of 179NC75 family members was affected by the N276D substitution and found that it had a profound impact (Fig 3B, lower left panel). In contrast, the N276D change had no effect on VRC01 binding, as previously reported [37] (Fig 3B). When monomeric gp120s from both YU2 and the clade A/E virus 93TH057 [38] were treated with EndoH, a glycosidase that removes N-linked oligomannose glycans, the binding of 179NC75 and its clonal variants was completely abolished (Fig 3C). To further probe the nature of the glycan-dependency of 179NC75, we tested binding of the Fab to BG505 SOSIP.664 trimers, (fully glycosylated, cleaved, native-like, soluble trimers [25]) produced in HEK293-6E cells in the presence and in the absence of the mannosidase I inhibitor kifunensine. HEK293-6E cells fully process glycans resulting in a mixture of complex-type and high-mannose N-glycans, while HEK293-6E cells treated with kifunensine, produce protein containing only high-mannose N-glycans. We observed that 179NC75 binds to BG505 SOSIP.664 trimer with processed glycans with a KD of ~90 nM, (S3 Fig) but cannot bind to trimers containing only high mannose glycans (S3 Fig, S6 Table). Hence, we conclude that 179NC75 is a glycan-dependent antibody that binds to the CD4bs in a way that involves the Asn276 residue and depends on the presence of complex glycans. In these respects, its epitope is similar to that of the HJ16 CD4bs bNAb.
We compared the neutralization potencies of 179NC75 to the ones of HJ16 [18]. For the 53 Tier-2 viruses that were tested against both HJ16 and 179NC75, 179NC75 neutralized more viruses than HJ16 (26 compared to 19), and was 20-fold more potent (IC50 of 0.118 μg/ml compared to 2.326 μg/ml) (Fig 3D).
Predicted germline variant of 179NC75 binds to BG505 SOSIP
Previous reports show that neutralizing antibodies bind BG505 SOSIP.664 trimers with higher affinity as opposed to non-neutralizing antibodies [25]. Therefore, as expected, the more potent variants of the 179NC75 clone, 179NC75 and 179NC1055, bound strongly to BG505 SOSIP.664-D7324 trimers in capture ELISA, while 179NC65 and 179NC21 bound weakly or not at all, respectively (S4 Fig).
Most predicted germline versions of CD4bs antibodies are unable to bind Env antigens [7]. To test whether the germline version of 179NC75 could bind the BG505 SOSIP.664-D7324 trimers, and assess the role of CDRH3 in trimer binding, we generated a germline version of 179NC75 (179NC75gl). The predicted germline version of the antibody was made as previously described by reverting the V and J segments of the heavy and light chains to their predicted germline sequences, while retaining the CDRH3 sequence as found in the mutated antibody [7,39,40]. For comparison, we used the previously published predicted germline versions of VRC01 [39,40], 3BNC60 [7], 1NC9 [7], CH103 [19] and HJ16 (constructed in the course of this study). Although all of the above mature CD4bs bNAbs bound the BG505 SOSIP.664-D7324 trimers, the only predicted germline antibody able to do so was 179NC75gl (Fig 4). An implication is that 179NC75 binding principally involves contacts made by the CDR3s, particularly the exceptionally long (24-residue) heavy chain CDR3.
Fig. 4. Mature versus predicted germline antibody binding to BG505 SOSIP.664-D7324 trimers in ELISA. 
(A) and (B) Binding of mutated CD4bs bNAbs (“MATURE”) and their reverted germline versions (“GERMLINE”) to BG505 SOSIP,664-D7324 trimers. The MGO.053 antibody [48] was used as a negative control. In vivo activity
The loop binding, glycan-dependent CD4bs bNAbs have not been tested for their activity in vivo. To address this issue, we treated six HIV-1YU2–infected hu-mice with 179NC75 for 5 weeks [8,29]. Monotherapy with 179NC75 resembled monotherapy with other bNAbs, in that there was a transient decrease in viral load in most of the treated animals followed by a rapid rebound [8,29,41] (Fig 5A and 5B). Viral env genes were cloned and sequenced from the day-28 plasma of 179NC75-treated mice, a time point where viremia had universally rebounded to levels similar to the day-0 value. Two types of mutations were consistently observed, both proximal to the CD4bs: the first eliminated the glycan-site at position N276; the second involved residues G459 or K460 (Fig 5C). In total, 13 sequences had only a mutation affecting the N276 glycan site, whereas 8 contained mutations in the region near position 460 and 7 sequences contained mutations in both regions (Fig 5C and 5D). In all mice the rebounding viruses carried at least one of these mutations. In mouse 1107 mutations in both areas were observed, resulting in the loss of the N276 glycan but the introduction of a potential N-linked glycosylation site (PNGS) at position 460 (Fig 5C). To confirm that the most commonly observed mutations did confer resistance, HIV-1YU2 Env-pseudoviruses containing one or both of the N276D and K460N substitutions were tested for their sensitivity to 179NC75. All three of the virus mutants were found to be 179NC75-resistant (Fig 5E). We also tested the HIVYU2 N280Y, N332K, N160K and G459D virus mutants. As expected, and consistent with the ELISA data, the N280Y substitution conferred complete resistance to 179NC75, while the N332K and N160K changes had no effect. The G459D mutant was also 179NC75-sensitive (Fig 5E). We conclude, that 179NC75 is a potent neutralizing antibody that exerts selection pressure on HIV-1YU2 in vivo and drives the emergence of resistant viruses with sequence changes proximal to the CD4bs.
Fig. 5. Hu-mice treatment with 179NC75. 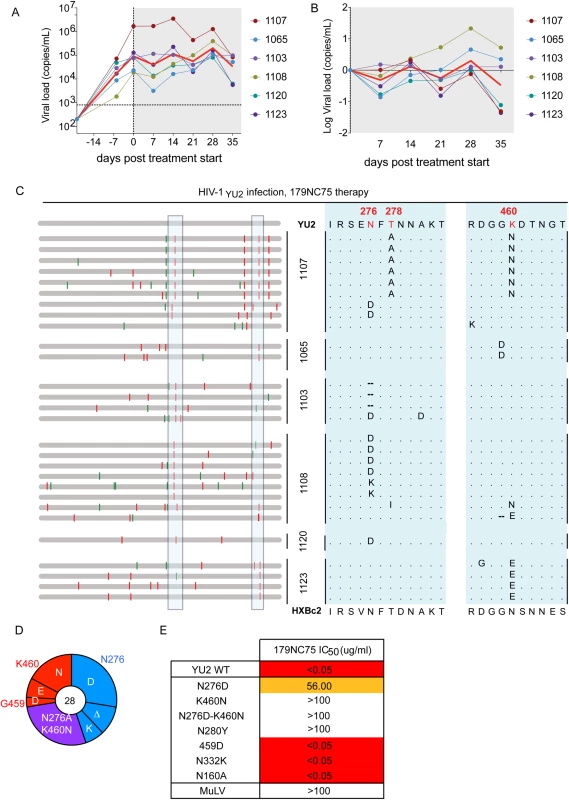
(A) Viral loads in six HIV-1YU2-infected hu-mice when 179NC75 was administered two weeks post-infection (day-0). The red line indicates the geometric mean value. (B) Log change in viral loads in each mouse compared to the day 0 value. (C) Left panel: each horizontal gray bar represents the sequence of a single gp120 clone aligned to HIV-1YU2. Synonymous and non-synonymous nucleotide substitutions are indicated in green and red, respectively. Sequences are grouped by the mouse from which they were obtained (center), indicated by the vertical black bars. An expanded view of the boxed areas is shown in the right panel. (D) Pie chart showing the recurrent mutations in gp120 compared to the wild-type HIV-1YU2 sequence. The number in the center of the pie denotes the total number of sequences cloned; the blue, red and purple slices represent the most consistently mutated areas in gp120, i.e., around residues 276 and 460, respectively. The sizes of the slices are proportional to the number of sequences that carried mutations. The original residue, as found in HIV-1YU2, is indicated on the outside of the pie chart, the various mutations in the cloned sequences are shown on the inside of the chart. “Δ” denotes a deletion of the residue. The purple slice indicates the presence of both the blue (276-area) and red (460-area) mutations. (E) The table shows IC50 values for 179NC75 neutralization of the wild-type HIV-1YU2 pseudovirus and mutants containing either the gp120 changes most frequently observed after treatment or other mutations that serve as comparators. Autologous viral selection by 179NC75
To test whether 179NC75 exerted selective pressure on the autologous virus found in subject EB179, we cloned env genes from the donor’s T cells obtained at the time of the leukapheresis. All nine gp120 sequences obtained contained Asn at position 460, introducing a PNGS at that position in eight of the nine sequences (Fig 6A–6C). Five sequences contained an Asn-Gly-Thr insertion immediately N-terminal to position N460, resulting in the sequence NGTNET, and therefore adding another PNGS to the one that was already at position 460. Five other sequences contained the N276S mutation, eliminating the PNGS at position 276. One of the nine sequences included both the Asn-Gly-Thr insertion at position 460 and the N276S change (Fig 6C). Of note is that this pattern of sequence changes is highly similar to the escape mutations seen in the env genes of the 179NC75-treated, HIV-1YU2-infected hu-mice (Fig 5). To test whether the autologous virus from patient EB179 is resistant to 179NC75, we cultured the donor’s CD4+ T cells from the same leukapheresis sample that was used for the antibody isolation. Outgrown virus was then tested for neutralization in the TZM.bl assay for neutralization by the EB179 polyclonal IgG (from the same time point), as well as by 179NC75 and other known bNAbs including the CD4bs antibody 3BNC117 [7], the V3-stem binding antibody 10–1074 [42] and the V1/V2 apex-binding antibody PG16 [43] (Fig 6D). As expected, the EB179 polyclonal IgG failed to neutralize the autologous virus. Amongst the two CD4bs antibodies, the autologous virus was fourfold more resistant to 179NC75 than to 3BNC117, suggesting that the EB179 antibody repertoire has CD4bs antibodies that differ from 3BNC117 and VRC01-class bNAbs. Interestingly, the autologous virus was also resistant to PG16 and 10–1074, indicating that the patient may have additional neutralizing antibodies bearing similar specificities in his antibody repertoire. Taken together, the data imply that loop-based, glycan-dependent CD4bs bNAbs of the 179NC75 family exert selective pressure on HIV-1 in vivo.
Fig. 6. Autologous viruses from EB179. 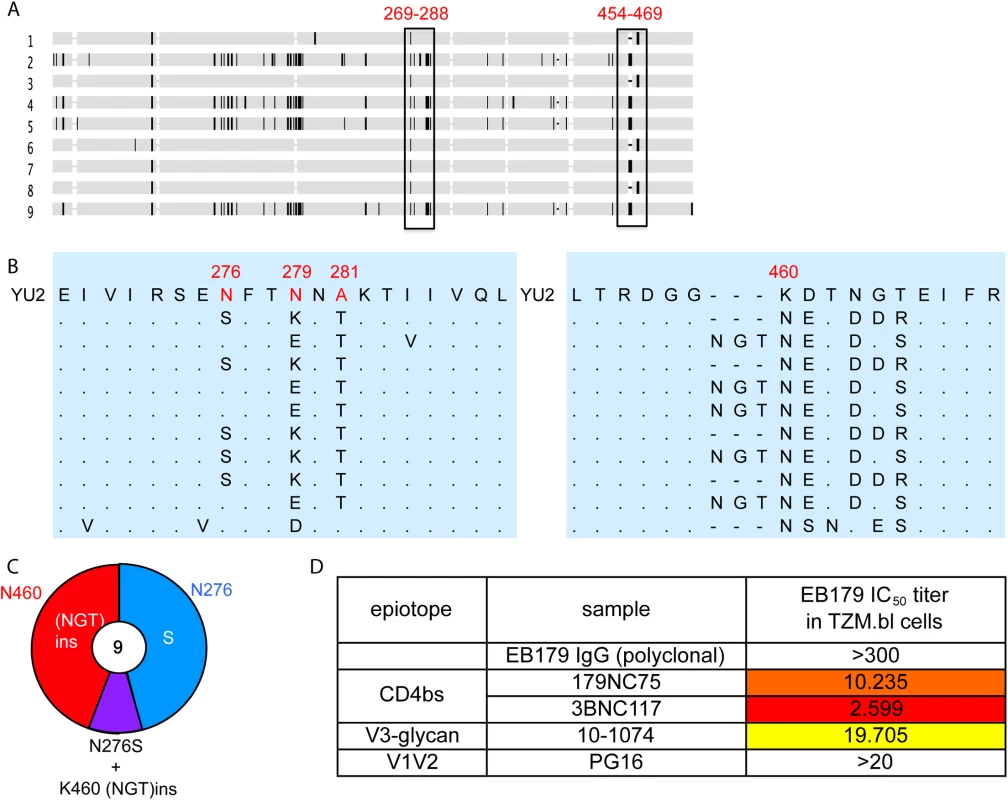
(A) Horizontal gray bars represents single gp120 sequences amplified and cloned from patient sera. The vertical rectangles show areas where recurrent mutations were found. (B) Expanded view of the most consistently mutated areas. (C) Pie chart comparing the recurrent mutations in gp120s from the EB179 autologous virus and HIV-1YU2. The number in the center of the pie denotes the total number of sequences cloned, and the sizes of slices are proportional to the number of sequences that carried the mutations; the color coding is same as used in Fig 5D. (D) The table shows IC50 values for the neutralization of the EB179 autologous culture virus by the autologous patient polyclonal IgG, as well as 179NC75, 3BNC117, 10–1074 and PG16 bNAbs. Discussion
The CD4bs is a highly conserved epitope on the HIV-1 Env and an important potential target for neutralizing antibodies. Although this site evolved to avoid antibody accessibility, two major groups of CD4bs bNAbs have been discovered [15]. The first group, exemplified by VRC01, is VH-restricted, IGVH1-2 or IGVH1-46, with the heavy chains positioned in a CD4-like orientation and CDRH2 making significant contacts with gp120 [6,7,15]. The CDRL3 [7,21,44], and in some cases also CDRL1 [6], of the corresponding light chains have to be short and compact to minimize potential interference and clashes with the glycans that surround the CD4bs. The emergence of these antibodies involves many somatic hypermutations, some of which are in the framework regions [45]. The second group of CD4bs bNAbs, which includes b12 and HJ16, is far more heterogeneous. These antibodies bind to gp120 via a CDRH3-dominated, loop based mechanism [15]. As might be expected, members of this group of CD4bs bNAbs arise from different VH segments and carry fewer somatic mutations [17–19]. The new antibody described in this study, 179NC75, is a loop binder that is closely related to HJ16. Similarly to HJ16, its Env-binding and virus-neutralizing activities are dependent on the N276 glycan [36]. Consistent with the CDRH3 loop-based mechanism of recognition that was described for antibodies that are not VH-restricted [15], when we generated the predicted germline version of 179NC75, where all mutations were reverted but the CDRH3 was retained, the antibody bound to BG505 SOSIP.664 trimers. This could indicate that any residual mutations present in the CDR3s of the reverted antibody might allow binding. Interestingly, the germline version of HJ16 also had some binding to BG505 SOSIP.664 trimers (Fig 4), however this binding was lower that the one of 179NC75, which could be attributed to a shorter CDRH3 (19 versus 24 residues).
Serum antibodies that are CD4bs-specific and N276-dependent have been described in HIV-1-infected individuals in two separate studies [32,46]. In the first study, an HJ16-type of CD4bs antibody response was found to be part of the second wave of serum neutralization in the CAP257 patient [46]. Viruses cloned from CAP257 after the emergence of these CD4bs antibodies carried an N276D or T278A mutation that were considered to be responses to antibody selection pressure [46]. In a second study, serum from individual VC1004 contained CD4bs-targeted NAbs that were sensitive to the N276D substitution but not D368R [47]. However, as the antibodies responsible for the serum activity were not cloned in either study much of what we know about the in vivo activity of these N276-dependent class of CD4bs antibodies is inferential. Our 179NC75 therapy experiments in HIV-1–infected hu-mice demonstrate that escape variants contain very similar, and sometimes identical, mutations to ones present in the autologous virus isolated from the infected human from whom the 179NC75 antibody was also derived. We conclude that the CDRH3-dominted N276-dependent CD4bs antibodies are effective at suppressing viremia in vivo and thence driving the emergence of escape variants.
Supporting Information
Zdroje
1. Temin HM (1993) Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci U S A 90 : 6900–6903. 7688465
2. McClure MO, Sattentau QJ, Beverley PC, Hearn JP, Fitzgerald AK, et al. (1987) HIV infection of primate lymphocytes and conservation of the CD4 receptor. Nature 330 : 487–489. 2446142
3. Scheid JF, Mouquet H, Feldhahn N, Walker BD, Pereyra F, et al. (2009) A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods 343 : 65–67. doi: 10.1016/j.jim.2008.11.012 19100741
4. Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, et al. (2009) Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458 : 636–640. doi: 10.1038/nature07930 19287373
5. Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, et al. (2010) Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329 : 856–861. doi: 10.1126/science.1187659 20616233
6. Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, et al. (2010) Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329 : 811–817. doi: 10.1126/science.1192819 20616231
7. Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, et al. (2011) Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333 : 1633–1637. doi: 10.1126/science.1207227 21764753
8. Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, et al. (2012) HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492 : 118–122. doi: 10.1038/nature11604 23103874
9. Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, et al. (2013) Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature 503 : 277–280. doi: 10.1038/nature12746 24172896
10. Shingai M, Donau OK, Plishka RJ, Buckler-White A, Mascola JR, et al. (2014) Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med.
11. Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, et al. (2013) Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature 503 : 224–228. doi: 10.1038/nature12744 24172905
12. Caskey M, Klein F, Lorenzi JC, Seaman MS, West AP Jr., et al. (2015) Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature.
13. Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, et al. (2011) Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333 : 1593–1602. doi: 10.1126/science.1207532 21835983
14. Zhou T, Zhu J, Wu X, Moquin S, Zhang B, et al. (2013) Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39 : 245–258. doi: 10.1016/j.immuni.2013.04.012 23911655
15. Zhou T, Lynch RM, Chen L, Acharya P, Wu X, et al. (2015) Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell.
16. West AP Jr., Diskin R, Nussenzweig MC, Bjorkman PJ (2012) Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci U S A 109: E2083–2090. doi: 10.1073/pnas.1208984109 22745174
17. Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, et al. (1994) Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266 : 1024–1027. 7973652
18. Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, et al. (2010) Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One 5: e8805. doi: 10.1371/journal.pone.0008805 20098712
19. Liao HX, Lynch R, Zhou T, Gao F, Alam SM, et al. (2013) Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496 : 469–476. doi: 10.1038/nature12053 23552890
20. Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, et al. (2007) Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445 : 732–737. 17301785
21. West AP Jr., Scharf L, Scheid JF, Klein F, Bjorkman PJ, et al. (2014) Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell 156 : 633–648. doi: 10.1016/j.cell.2014.01.052 24529371
22. Dey B, Svehla K, Xu L, Wycuff D, Zhou T, et al. (2009) Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog 5: e1000445. doi: 10.1371/journal.ppat.1000445 19478876
23. Freund NT, Scheid JF, Mouquet H, Nussenzweig MC (2015) Amplification of highly mutated human Ig lambda light chains from an HIV-1 infected patient. J Immunol Methods.
24. Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, et al. (2008) Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 329 : 112–124. 17996249
25. Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, et al. (2013) A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9: e1003618. doi: 10.1371/journal.ppat.1003618 24068931
26. Sok D, van Gils MJ, Pauthner M, Julien JP, Saye-Francisco KL, et al. (2014) Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc Natl Acad Sci U S A 111 : 17624–17629. doi: 10.1073/pnas.1415789111 25422458
27. Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, et al. (2005) Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79 : 10108–10125. 16051804
28. van 't Wout AB, Schuitemaker H, Kootstra NA (2008) Isolation and propagation of HIV-1 on peripheral blood mononuclear cells. Nat Protoc 3 : 363–370. doi: 10.1038/nprot.2008.3 18323807
29. Horwitz JA, Halper-Stromberg A, Mouquet H, Gitlin AD, Tretiakova A, et al. (2013) HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc Natl Acad Sci U S A 110 : 16538–16543. doi: 10.1073/pnas.1315295110 24043801
30. Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, et al. (2014) Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509 : 55–62.
31. Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, et al. (2012) Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med 18 : 1688–1692. doi: 10.1038/nm.2985 23086475
32. Sather DN, Carbonetti S, Malherbe D, Pissani F, Stuart AB, et al. (2014) Emergence of broadly neutralizing antibodies and viral co-evolution in two subjects during the early stages of infection with the human immunodeficiency virus type 1. J Virol.
33. Yang X, Farzan M, Wyatt R, Sodroski J (2000) Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J Virol 74 : 5716–5725. 10823881
34. Sellhorn G, Kraft Z, Caldwell Z, Ellingson K, Mineart C, et al. (2012) Engineering, expression, purification, and characterization of stable clade A/B recombinant soluble heterotrimeric gp140 proteins. J Virol 86 : 128–142. doi: 10.1128/JVI.06363-11 22031951
35. Scharf L, Scheid JF, Lee JH, West AP Jr., Chen C, et al. (2014) Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Rep 7 : 785–795. doi: 10.1016/j.celrep.2014.04.001 24767986
36. Balla-Jhagjhoorsingh SS, Corti D, Heyndrickx L, Willems E, Vereecken K, et al. (2013) The N276 glycosylation site is required for HIV-1 neutralization by the CD4 binding site specific HJ16 monoclonal antibody. PLoS One 8: e68863. doi: 10.1371/journal.pone.0068863 23874792
37. Li Y, O'Dell S, Walker LM, Wu X, Guenaga J, et al. (2011) Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J Virol 85 : 8954–8967. doi: 10.1128/JVI.00754-11 21715490
38. Anderson JP, Rodrigo AG, Learn GH, Madan A, Delahunty C, et al. (2000) Testing the hypothesis of a recombinant origin of human immunodeficiency virus type 1 subtype E. J Virol 74 : 10752–10765. 11044120
39. McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, et al. (2013) Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med 210 : 655–663. doi: 10.1084/jem.20122824 23530120
40. Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, et al. (2013) Rational HIV immunogen design to target specific germline B cell receptors. Science 340 : 711–716. doi: 10.1126/science.1234150 23539181
41. Halper-Stromberg A, Lu CL, Klein F, Horwitz JA, Bournazos S, et al. (2014) Broadly Neutralizing Antibodies and Viral Inducers Decrease Rebound from HIV-1 Latent Reservoirs in Humanized Mice. Cell 158 : 989–999. doi: 10.1016/j.cell.2014.07.043 25131989
42. Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, et al. (2012) Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 109: E3268–3277. doi: 10.1073/pnas.1217207109 23115339
43. Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, et al. (2013) Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol 20 : 804–813. doi: 10.1038/nsmb.2600 23708607
44. Scharf L, West AP Jr., Gao H, Lee T, Scheid JF, et al. (2013) Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody. Proc Natl Acad Sci U S A 110 : 6049–6054. doi: 10.1073/pnas.1303682110 23524883
45. Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, et al. (2013) Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153 : 126–138. doi: 10.1016/j.cell.2013.03.018 23540694
46. Wibmer CK, Bhiman JN, Gray ES, Tumba N, Abdool Karim SS, et al. (2013) Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog 9: e1003738. doi: 10.1371/journal.ppat.1003738 24204277
47. Sather DN, Carbonetti S, Malherbe D, Pissani F, Stuart AB, et al. (2014) Development of Broadly Neutralizing Anti-HIV-1 Antibodies during Natural Infection through Early Epitope Acquisition and Subsequent Maturation. AIDS Res Hum Retroviruses 30 Suppl 1: A35.
48. Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, et al. (2010) Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature 467 : 591–595. doi: 10.1038/nature09385 20882016
49. Balla-Jhagjhoorsingh SS, Willems B, Heyndrickx L, Heyndrickx L, Vereecken K, et al. (2011) Characterization of neutralizing profiles in HIV-1 infected patients from whom the HJ16, HGN194 and HK20 mAbs were obtained. PLoS One 6: e25488. doi: 10.1371/journal.pone.0025488 22016769
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized VirusČlánek Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell ProliferationČlánek Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite InterfaceČlánek Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 10- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Expression of Concern: Misregulation of Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis
- Preparing for the Next Epidemic with Basic Virology
- Effectively Communicating the Uncertainties Surrounding Ebola Virus Transmission
- Translating Basic Research into Clinical Applications: Malaria Research at an NIH Lab
- A Gut Odyssey: The Impact of the Microbiota on Spore Formation and Germination
- Papillomavirus E6 Oncoproteins Take Common Structural Approaches to Solve Different Biological Problems
- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Dimensions of Horizontal Gene Transfer in Eukaryotic Microbial Pathogens
- Addressing the Complications of Ebola and Other Viral Hemorrhagic Fever Infections: Using Insights from Bacterial and Fungal Sepsis
- Time for Chocolate: Current Understanding and New Perspectives on Cacao Witches’ Broom Disease Research
- Ganglioside and Non-ganglioside Mediated Host Responses to the Mouse Polyomavirus
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Structure Elucidation of Coxsackievirus A16 in Complex with GPP3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses
- CD39 Expression Identifies Terminally Exhausted CD8 T Cells
- Abiotic Stresses Antagonize the Rice Defence Pathway through the Tyrosine-Dephosphorylation of OsMPK6
- Dissociation of Tissue Destruction and Bacterial Expansion during Bubonic Plague
- Interferon-γ: The Jekyll and Hyde of Malaria
- CCR2 Inflammatory Dendritic Cells and Translocation of Antigen by Type III Secretion Are Required for the Exceptionally Large CD8 T Cell Response to the Protective YopE Epitope during Infection
- A New Glycan-Dependent CD4-Binding Site Neutralizing Antibody Exerts Pressure on HIV-1
- The Suramin Derivative NF449 Interacts with the 5-fold Vertex of the Enterovirus A71 Capsid to Prevent Virus Attachment to PSGL-1 and Heparan Sulfate
- Trans-generational Immune Priming Protects the Eggs Only against Gram-Positive Bacteria in the Mealworm Beetle
- Peripheral Vγ9Vδ2 T Cells Are a Novel Reservoir of Latent HIV Infection
- Respiratory Syncytial Virus Disease Is Mediated by Age-Variable IL-33
- TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized Virus
- Modeling the Effects of Vorinostat Reveals both Transient and Delayed HIV Transcriptional Activation and Minimal Killing of Latently Infected Cells
- Identification of a Novel Lipoprotein Regulator of Spore Germination
- Calcium Regulation of Hemorrhagic Fever Virus Budding: Mechanistic Implications for Host-Oriented Therapeutic Intervention
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
- Comparative Life Cycle Transcriptomics Revises Genome Annotation and Links a Chromosome Duplication with Parasitism of Vertebrates
- The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy
- Carcinogenic Parasite Secretes Growth Factor That Accelerates Wound Healing and Potentially Promotes Neoplasia
- Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell Proliferation
- Dengue Virus Infection of Requires a Putative Cysteine Rich Venom Protein
- Distinct Viral and Mutational Spectrum of Endemic Burkitt Lymphoma
- Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite Interface
- Phenotypic and Functional Alterations in Circulating Memory CD8 T Cells with Time after Primary Infection
- Systematic Identification of Cyclic-di-GMP Binding Proteins in Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems
- Influenza Transmission in the Mother-Infant Dyad Leads to Severe Disease, Mammary Gland Infection, and Pathogenesis by Regulating Host Responses
- Myeloid Cell Arg1 Inhibits Control of Arthritogenic Alphavirus Infection by Suppressing Antiviral T Cells
- The White-Nose Syndrome Transcriptome: Activation of Anti-fungal Host Responses in Wing Tissue of Hibernating Little Brown Myotis
- Influenza Virus Reassortment Is Enhanced by Semi-infectious Particles but Can Be Suppressed by Defective Interfering Particles
- Identification of the Mechanisms Causing Reversion to Virulence in an Attenuated SARS-CoV for the Design of a Genetically Stable Vaccine
- Differentiation-Dependent KLF4 Expression Promotes Lytic Epstein-Barr Virus Infection in Epithelial Cells
- The Histone Acetyltransferase Hat1 Regulates Stress Resistance and Virulence via Distinct Chromatin Assembly Pathways
- C-di-GMP Regulates Motile to Sessile Transition by Modulating MshA Pili Biogenesis and Near-Surface Motility Behavior in
- Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
- Crystal Structure of the Human Cytomegalovirus Glycoprotein B
- Depletion of . GlmU from Infected Murine Lungs Effects the Clearance of the Pathogen
- Immunologic Control of Papillomavirus Type 1
- Requires Host Rab1b for Survival in Macrophages
- Structure Analysis Uncovers a Highly Diverse but Structurally Conserved Effector Family in Phytopathogenic Fungi
- PD-L1 Expression on Retrovirus-Infected Cells Mediates Immune Escape from CD8 T Cell Killing
- Phospho-dependent Regulation of SAMHD1 Oligomerisation Couples Catalysis and Restriction
- IL-4 Induced Innate CD8 T Cells Control Persistent Viral Infection
- Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
- BCG Skin Infection Triggers IL-1R-MyD88-Dependent Migration of EpCAM CD11b Skin Dendritic cells to Draining Lymph Node During CD4+ T-Cell Priming
- Antigenic Characterization of the HCMV gH/gL/gO and Pentamer Cell Entry Complexes Reveals Binding Sites for Potently Neutralizing Human Antibodies
- Rescue of a Plant Negative-Strand RNA Virus from Cloned cDNA: Insights into Enveloped Plant Virus Movement and Morphogenesis
- Geminivirus Activates to Accelerate Cytoplasmic DCP2-Mediated mRNA Turnover and Weakens RNA Silencing in
- Disruption of Sphingolipid Biosynthesis Blocks Phagocytosis of
- The Fungal Exopolysaccharide Galactosaminogalactan Mediates Virulence by Enhancing Resistance to Neutrophil Extracellular Traps
- The Timing of Stimulation and IL-2 Signaling Regulate Secondary CD8 T Cell Responses
- Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity
- The Dual Role of an ESCRT-0 Component HGS in HBV Transcription and Naked Capsid Secretion
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Interferon-γ: The Jekyll and Hyde of Malaria
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



