-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSTrengthening the REporting of Genetic Association Studies (STREGA)— An Extension of the STROBE Statement
article has not abstract
Published in the journal: . PLoS Med 6(2): e32767. doi:10.1371/journal.pmed.1000022
Category: Guidelines and Guidance
doi: https://doi.org/10.1371/journal.pmed.1000022Summary
article has not abstract
The rapidly evolving evidence on genetic associations is crucial to integrating human genomics into the practice of medicine and public health [1,2]. Genetic factors are likely to affect the occurrence of numerous common diseases, and therefore identifying and characterizing the associated risk (or protection) will be important in improving the understanding of etiology and potentially for developing interventions based on genetic information. The number of publications on the associations between genes and diseases has increased tremendously; with more than 34 000 published articles, the annual number has more than doubled between 2001 and 2008 [3,4]. Articles on genetic associations have been published in about 1500 journals and in several languages.
Despite the many similarities between genetic association studies and “classical” observational epidemiologic studies (that is, cross-sectional, case-control, and cohort) of lifestyle and environmental factors, genetic association studies present several specific challenges including an unprecedented volume of new data [5,6] and the likelihood of very small individual effects. Genes may operate in complex pathways with gene-environment and gene-gene interactions [7]. Moreover, the current evidence base on gene-disease associations is fraught with methodological problems [8–10]. Inadequate reporting of results, even from well-conducted studies, hampers assessment of a study's strengths and weaknesses, and hence the integration of evidence [11].
Summary
Making sense of rapidly evolving evidence on genetic associations is crucial to making genuine advances in human genomics and the eventual integration of this information in the practice of medicine and public health. Assessment of the strengths and weaknesses of this evidence, and hence the ability to synthesize it, has been limited by inadequate reporting of results. The STrengthening the REporting of Genetic Association studies (STREGA) initiative builds on the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement and provides additions to 12 of the 22 items on the STROBE checklist. The additions concern population stratification, genotyping errors, modelling haplotype variation, Hardy-Weinberg equilibrium, replication, selection of participants, rationale for choice of genes and variants, treatment effects in studying quantitative traits, statistical methods, relatedness, reporting of descriptive and outcome data, and the volume of data issues that are important to consider in genetic association studies. The STREGA recommendations do not prescribe or dictate how a genetic association study should be designed but seek to enhance the transparency of its reporting, regardless of choices made during design, conduct, or analysis.
Although several commentaries on the conduct, appraisal and/or reporting of genetic association studies have so far been published [12–39], their recommendations differ. For example, some papers suggest that replication of findings should be part of the publication [12,13,16,17,23,26,34–36] whereas others consider this suggestion unnecessary or even unreasonable [21,40–44]. In many publications, the guidance has focused on genetic association studies of specific diseases [14,15,17,19,22,23,25,26,31–38] or the design and conduct of genetic association studies [13–15,17,19,20,22,23,25,30–32,35,36] rather than on the quality of the reporting.
Despite increasing recognition of these problems, the quality of reporting genetic association studies needs to be improved [45–49]. For example, an assessment of a random sample of 315 genetic association studies published from 2001 to 2003 found that most studies provided some qualitative descriptions of the study participants (for example, origin and enrolment criteria), but reporting of quantitative descriptors, such as age and sex, was variable [49]. In addition, completeness of reporting of methods that allow readers to assess potential biases (for example, number of exclusions or number of samples that could not be genotyped) varied [49]. Only some studies described methods to validate genotyping or mentioned whether research staff were blinded to outcome. The same problems persisted in a smaller sample of studies published in 2006 [49]. Lack of transparency and incomplete reporting have raised concerns in a range of health research fields [11,50–53] and poor reporting has been associated with biased estimates of effects in clinical intervention studies [54].
The main goal of this article is to propose and justify a set of guiding principles for reporting results of genetic association studies. The epidemiology community has recently developed the Strengthening the Reporting of Observational studies in Epidemiology (STROBE) Statement for cross-sectional, case-control, and cohort studies [55,56]. Given the relevance of general epidemiologic principles for genetic association studies, we propose recommendations in an extension of the STROBE Statement called the STrengthening the REporting of Genetic Association studies (STREGA) Statement. The recommendations of the STROBE Statement have a strong foundation because they are based on empirical evidence on the reporting of observational studies, and they involved extensive consultations in the epidemiologic research community [56]. We have sought to identify gaps and areas of controversy in the evidence regarding potential biases in genetic association studies. With the recommendations, we have indicated available empirical or theoretical work that has demonstrated or suggested that a methodological feature of a study can influence the direction or magnitude of the association observed. We acknowledge that for many items, no such evidence exists. The intended audience for the reporting guideline is broad and includes epidemiologists, geneticists, statisticians, clinician scientists, and laboratory-based investigators who undertake genetic association studies. In addition, it includes “users” of such studies who wish to understand the basic premise, design, and limitations of genetic association studies in order to interpret the results. The field of genetic associations is evolving very rapidly with the advent of genome-wide association investigations, high-throughput platforms assessing genetic variability beyond common single nucleotide polymorphisms (SNPs) (for example, copy number variants, rare variants), and eventually routine full sequencing of samples from large populations. Our recommendations are not intended to support or oppose the choice of any particular study design or method. Instead, they are intended to maximize the transparency, quality and completeness of reporting of what was done and found in a particular study.
Methods
A multidisciplinary group developed the STREGA Statement by using literature review, workshop presentations and discussion, and iterative electronic correspondence after the workshop. Thirty-three of 74 invitees participated in the STREGA workshop in Ottawa, Ontario, Canada, in June, 2006. Participants included epidemiologists, geneticists, statisticians, journal editors and graduate students.
Before the workshop, an electronic search was performed to identify existing reporting guidance for genetic association studies. Workshop participants were also asked to identify any additional guidance. They prepared brief presentations on existing reporting guidelines, empirical evidence on reporting of genetic association studies, the development of the STROBE Statement, and several key areas for discussion that were identified on the basis of consultations before the workshop. These areas included the selection and participation of study participants, rationale for choice of genes and variants investigated, genotyping errors, methods for inferring haplotypes, population stratification, assessment of Hardy-Weinberg equilibrium (HWE), multiple testing, reporting of quantitative (continuous) outcomes, selectively reporting study results, joint effects and inference of causation in single studies. Additional resources to inform workshop participants were the HuGENet handbook [57,58], examples of data extraction forms from systematic reviews or meta-analyses, articles on guideline development [59,60] and the checklists developed for STROBE. To harmonize our recommendations for genetic association studies with those for observational epidemiologic studies, we communicated with the STROBE group during the development process and sought their comments on the STREGA draft documents. We also provided comments on the developing STROBE Statement and its associated explanation and elaboration document [56].
Results
In Table 1, we present the STREGA recommendations, an extension to the STROBE checklist [55] for genetic association studies (an editable version of Table 1 is provided as Table S1 under Supporting Information). The resulting STREGA checklist provides additions to 12 of the 22 items on the STROBE checklist. During the workshop and subsequent consultations, we identified five main areas of special interest that are specific to, or especially relevant in, genetic association studies: genotyping errors, population stratification, modelling haplotype variation, HWE and replication. We elaborate on each of these areas, starting each section with the corresponding STREGA recommendation, followed by a brief outline of the issue and an explanation for the recommendations. Complementary information on these areas and the rationale for additional STREGA recommendations relating to selection of participants, choice of genes and variants selected, treatment effects in studying quantitative traits, statistical methods, relatedness, reporting of descriptive and outcome data, and issues of data volume, are presented in Table 2.
Tab. 1. STREGA Reporting Recommendations, Extended from STROBE Statement 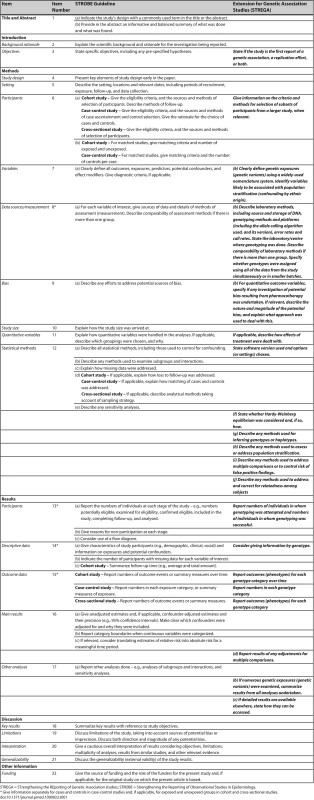
STREGA = STrengthening the REporting of Genetic Association studies; STROBE = Strengthening the Reporting of Observational Studies in Epidemiology. Tab. 2. Rationale for Inclusion of Topics in the STREGA Recommendations 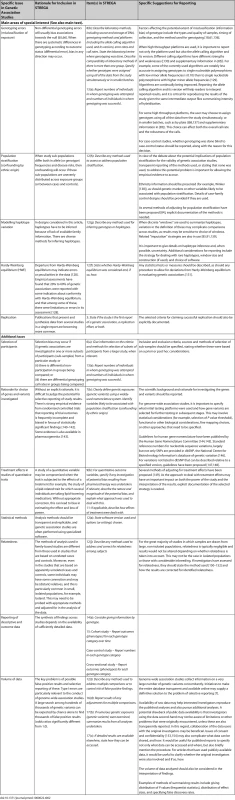
Genotyping Errors
Recommendation for reporting of methods (Table 1, item 8(b)): Describe laboratory methods, including source and storage of DNA, genotyping methods and platforms (including the allele calling algorithm used, and its version), error rates and call rates. State the laboratory/centre where genotyping was done. Describe comparability of laboratory methods if there is more than one group. Specify whether genotypes were assigned using all of the data from the study simultaneously or in smaller batches.
Recommendation for reporting of results (Table 1, item 13(a)): Report numbers of individuals in whom genotyping was attempted and numbers of individuals in whom genotyping was successful.
Genotyping errors can occur as a result of effects of the DNA sequence flanking the marker of interest, poor quality or quantity of the DNA extracted from biological samples, biochemical artefacts, poor equipment precision or equipment failure, or human error in sample handling, conduct of the array or handling the data obtained from the array [61]. A commentary published in 2005 on the possible causes and consequences of genotyping errors observed that an increasing number of researchers were aware of the problem, but that the effects of such errors had largely been neglected [61]. The magnitude of genotyping errors has been reported to vary between 0.5% and 30% [61–64]. In high-throughput centres, an error rate of 0.5% per genotype has been observed for blind duplicates that were run on the same gel [64]. This lower error rate reflects an explicit choice of markers for which genotyping rates have been found to be highly repeatable and whose individual polymerase chain reactions (PCR) have been optimized. Non-differential genotyping errors, that is, those that do not differ systematically according to outcome status, will usually bias associations towards the null [65,66], just as for other non-differential errors. The most marked bias occurs when genotyping sensitivity is poor and genotype prevalence is high (>85%) or, as the corollary, when genotyping specificity is poor and genotype prevalence is low (<15%) [65]. When measurement of the environmental exposure has substantial error, genotyping errors of the order of 3% can lead to substantial under-estimation of the magnitude of an interaction effect [67]. When there are systematic differences in genotyping according to outcome status (differential error), bias in any direction may occur. Unblinded assessment may lead to differential misclassification. For genome-wide association studies of SNPs, differential misclassification between comparison groups (for example, cases and controls) can occur because of differences in DNA storage, collection or processing protocols, even when the genotyping itself meets the highest possible standards [68]. In this situation, using samples blinded to comparison group to determine the parameters for allele calling could still lead to differential misclassification. To minimize such differential misclassification, it would be necessary to calibrate the software separately for each group. This is one of the reasons for our recommendation to specify whether genotypes were assigned using all of the data from the study simultaneously or in smaller batches.
Population Stratification
Recommendation for reporting of methods (Table 1, item 12(h)): Describe any methods used to assess or address population stratification.
Population stratification is the presence within a population of subgroups among which allele (or genotype; or haplotype) frequencies and disease risks differ. When the groups compared in the study differ in their proportions of the population subgroups, an association between the genotype and the disease being investigated may reflect the genotype being an indicator identifying a population subgroup rather than a causal variant. In this situation, population subgroup is a confounder because it is associated with both genotype frequency and disease risk. The potential implications of population stratification for the validity of genetic association studies have been debated [69–83]. Modelling the possible effect of population stratification (when no effort has been made to address it) suggests that the effect is likely to be small in most situations [75,76,78–80]. Meta-analyses of 43 gene-disease associations comprising 697 individual studies showed consistent associations across groups of different ethnic origin [80], and thus provide evidence against a large effect of population stratification, hidden or otherwise. However, as studies of association and interaction typically address moderate or small effects and hence require large sample sizes, a small bias arising from population stratification may be important [81]. Study design (case-family control studies) and statistical methods [84] have been proposed to address population stratification, but so far few studies have used these suggestions [49]. Most of the early genome-wide association studies used family-based designs or such methods as genomic control and principal components analysis [85,86] to control for stratification. These approaches are particularly appropriate for addressing bias when the identified genetic effects are very small (odds ratio <1.20), as has been the situation in many recent genome-wide association studies [85,87–105]. In view of the debate about the potential implications of population stratification for the validity of genetic association studies, we recommend transparent reporting of the methods used, or stating that none was used, to address this potential problem. This reporting will enable empirical evidence to accrue about the effects of population stratification and methods to address it.
Modelling Haplotype Variation
Recommendation for reporting of methods (Table 1, item 12(g)): Describe any methods used for inferring genotypes or haplotypes.
A haplotype is a combination of specific alleles at neighbouring genes that tends to be inherited together. There has been considerable interest in modelling haplotype variation within candidate genes. Typically, the number of haplotypes observed within a gene is much smaller than the theoretical number of all possible haplotypes [106,107]. Motivation for utilizing haplotypes comes, in large part, from the fact that multiple SNPs may “tag” an untyped variant more effectively than a single typed variant. The subset of SNPs used in such an approach is called “haplotype tagging” SNPs. Implicitly, an aim of haplotype tagging is to reduce the number of SNPs that have to be genotyped, while maintaining statistical power to detect an association with the phenotype. Maps of human genetic variation are becoming more complete, and large scale genotypic analysis is becoming increasingly feasible. In consequence, it is possible that modelling haplotype variation will become more focussed on rare causal variants, because these may not be included in the genotyping platforms.
In most current, large-scale genetic association studies, data are collected as unphased multilocus genotypes (that is, which alleles are aligned together on particular segments of chromosome is unknown). It is common in such studies to use statistical methods to estimate haplotypes [108–111], and their accuracy and efficiency have been discussed [112–116]. Some methods attempt to make use of a concept called haplotype “blocks” [117,118], but the results of these methods are sensitive to the specific definitions of the “blocks” [119,120]. Reporting of the methods used to infer individual haplotypes and population haplotype frequencies, along with their associated uncertainties should enhance our understanding of the possible effects of different methods of modelling haplotype variation on study results as well as enabling comparison and syntheses of results from different studies.
Information on common patterns of genetic variation revealed by the International Haplotype Map (HapMap) Project [107] can be applied in the analysis of genome-wide association studies to infer genotypic variation at markers not typed directly in these studies [121,122]. Essentially, these methods perform haplotype-based tests but make use of information on variation in a set of reference samples (for example, HapMap) to guide the specific tests of association, collapsing a potentially large number of haplotypes into two classes (the allelic variation) at each marker. It is expected that these techniques will increase power in individual studies, and will aid in combining data across studies, and even across differing genotyping platforms. If imputation procedures have been used, it is useful to know the method, accuracy thresholds for acceptable imputation, how imputed genotypes were handled or weighted in the analysis, and whether any associations based on imputed genotypes were also verified on the basis of direct genotyping at a subsequent stage.
Hardy-Weinberg Equilibrium
Recommendation for reporting of methods (Table 1, item 12(f)): State whether Hardy-Weinberg equilibrium was considered and, if so, how.
Hardy-Weinberg equilibrium has become widely accepted as an underlying model in population genetics after Hardy [123] and Weinberg [124] proposed the concept that genotype frequencies at a genetic locus are stable within one generation of random mating; the assumption of HWE is equivalent to the independence of two alleles at a locus. Views differ on whether testing for departure from HWE is a useful method to detect errors or peculiarities in the data set, and also the method of testing [125]. In particular, it has been suggested that deviation from HWE may be a sign of genotyping errors [126–128]. Testing for departure from HWE has a role in detecting gross errors of genotyping in large-scale genotyping projects such as identifying SNPs for which the clustering algorithms used to call genotypes have broken down [85,129]. However, the statistical power to detect less important errors of genotyping by testing for departure from HWE is low [130] and, in hypothetical data, the presence of HWE was generally not altered by the introduction of genotyping errors [131]. Furthermore, the assumptions underlying HWE, including random mating, lack of selection according to genotype, and absence of mutation or gene flow, are rarely met in human populations [132,133]. In five of 42 gene-disease associations assessed in meta-analyses of almost 600 studies, the results of studies that violated HWE significantly differed from results of studies that conformed to the model [134]. Moreover, the study suggested that exclusion of HWE-violating studies may result in loss of the statistical significance of some postulated gene-disease associations and that adjustment for the magnitude of deviation from the model may also have the same consequence for some other gene-disease associations. Given the differing views about the value of testing for departure from HWE and about the test methods, transparent reporting of whether such testing was done and, if so, the method used, is important for allowing the empirical evidence to accrue.
For massive-testing platforms, such as genome-wide association studies, it might be expected that many false-positive violations of HWE would occur if a lenient P value threshold were set. There is no consensus on the appropriate P value threshold for HWE-related quality control in this setting. So, we recommend that investigators state which threshold they have used, if any, to exclude specific polymorphisms from further consideration. For SNPs with low minor allele frequencies, substantially more significant results than expected by chance have been observed, and the distribution of alleles at these loci has often been found to show departure from HWE.
For genome-wide association studies, another approach that has been used to detect errors or peculiarities in the data set (due to population stratification, genotyping error, HWE deviations or other reasons) has been to construct quantile-quantile (Q/Q) plots whereby observed association statistics or calculated P values for each SNP are ranked in order from smallest to largest and plotted against the expected null distribution [129,130]. The shape of the curve can lend insight into whether or not systematic biases are present.
Replication
Recommendation: State if the study is the first report of a genetic association, a replication effort, or both. (Table 1, item 3)
Articles that present and synthesize data from several studies in a single report are becoming more common. In particular, many genome-wide association analyses describe several different study populations, sometimes with different study designs and genotyping platforms, and in various stages of discovery and replication [129,130]. When data from several studies are presented in a single original report, each of the constituent studies and the composite results should be fully described. For example, a discussion of sample size and the reason for arriving at that size would include clear differentiation between the initial group (those that were typed with the full set of SNPs) and those that were included in the replication phase only (typed with a reduced set of SNPs) [129,130]. Describing the methods and results in sufficient detail would require substantial space in print, but options for publishing additional information on the study online make this possible.
Discussion
The choices made for study design, conduct and data analysis potentially influence the magnitude and direction of results of genetic association studies. However, the empirical evidence on these effects is insufficient. Transparency of reporting is thus essential for developing a better evidence base (Table 2). Transparent reporting helps address gaps in empirical evidence [45], such as the effects of incomplete participation and genotyping errors. It will also help assess the impact of currently controversial issues such as population stratification, methods of inferring haplotypes, departure from HWE and multiple testing on effect estimates under different study conditions.
The STREGA Statement proposes a minimum checklist of items for reporting genetic association studies. The statement has several strengths. First, it is based on existing guidance on reporting observational studies (STROBE). Second, it was developed from discussions of an interdisciplinary group that included epidemiologists, geneticists, statisticians, journal editors, and graduate students, thus reflecting a broad collaborative approach in terminology accessible to scientists from diverse disciplines. Finally, it explicitly describes the rationale for the decisions (Table 2) and has a clear plan for dissemination and evaluation.
The STREGA recommendations are available at http://www.strega-statement.org/. We welcome comments, which will be used to refine future versions of the recommendations. We note that little is known about the most effective ways to apply reporting guidelines in practice, and that therefore it has been suggested that editors and authors collect, analyze, and report their experiences in using such guidelines [135]. We consider that the STREGA recommendations can be used by authors, peer reviewers and editors to improve the reporting of genetic association studies. We invite journals to endorse STREGA, for example by including STREGA and its Web address in their Instructions for Authors and by advising authors and peer reviewers to use the checklist as a guide. It has been suggested that reporting guidelines are most helpful if authors keep the general content of the guideline items in mind as they write their initial drafts, then refer to the details of individual items as they critically appraise what they have written during the revision process [135]. We emphasize that the STREGA reporting guidelines should not be used for screening submitted manuscripts to determine the quality or validity of the study being reported. Adherence to the recommendations may make some manuscripts longer, and this may be seen as a drawback in an era of limited space in a print journal. However, the ability to post information on the Web should alleviate this concern. The place in which supplementary information is presented can be decided by authors and editors of the individual journal.
We hope that the recommendations stimulate transparent and improved reporting of genetic association studies. In turn, better reporting of original studies would facilitate the synthesis of available research results and the further development of study methods in genetic epidemiology with the ultimate goal of improving the understanding of the role of genetic factors in the cause of diseases.
Supporting Information
Zdroje
1. KhouryMJLittleJBurkeW
2004
Human genome epidemiology: Scope and strategies.
KhouryMJLittleJBurkeW
editors In
Human genome epidemiology: A scientific foundation for using genetic information to improve health and prevent disease
New York
Oxford University Press
3
16
2. Genomics, Health and Society Working Group
2004
Genomics, health and society. Emerging issues for public policy
Ottawa
Government of Canada Policy Research Initiative
3. LinBKClyneMWalshMGomezOYuW
2006
Tracking the epidemiology of human genes in the literature: The HuGE published literature database.
Am J Epidemiol
164
1
4
4. YuWYesupriyaAClyneMWulfAGwinnM
2008
HuGE Literature Finder. HuGE Navigator.
Available: http://www.hugenavigator.net/HuGENavigator/searchSummary.do?firstQuery=Gene-disease+association&publitSearchType=now&whichContinue=firststart&check=n&dbType=publit&Mysubmit=go. Accessed 15 December 2008
5. LawrenceRWEvansDMCardonLR
2005
Prospects and pitfalls in whole genome association studies.
Philos Trans R Soc Lond B Biol Sci
360
1589
1595
6. ThomasDC
2006
Are we ready for genome-wide association studies?
Cancer Epidemiol Biomarkers Prev
15
595
598
7. KhouryMJLittleJGwinnMIoannidisJP
2007
On the synthesis and interpretation of consistent but weak gene-disease associations in the era of genome-wide association studies.
Int J Epidemiol
36
439
445
8. LittleJKhouryMJBradleyLClyneMGwinnM
2003
The human genome project is complete. How do we develop a handle for the pump?
Am J Epidemiol
157
667
673
9. IoannidisJPBernsteinJBoffettaPDaneshJDolanS
2005
A network of investigator networks in human genome epidemiology.
Am J Epidemiol
162
302
304
10. IoannidisJPGwinnMLittleJHigginsJPBernsteinJL
2006
A road map for efficient and reliable human genome epidemiology.
Nat Genet
38
3
5
11. von ElmEEggerM
2004
The scandal of poor epidemiological research.
BMJ
329
868
869
12. [Anonymous]
1999
Freely associating (editorial).
Nat Genet
22
1
2
13. CardonLBellJ
2001
Association study designs for complex diseases.
Nat Rev Genet
2
91
99
14. WeissS
2001
Association studies in asthma genetics.
Am J Respir Crit Care Med
164
2014
2015
15. WeissSTSilvermanEKPalmerLJ
2001
Case-control association studies in pharmacogenetics.
Pharmacogenomics J
1
157
158
16. CooperDNNussbaumRLKrawczakM
2002
Proposed guidelines for papers describing DNA polymorphism-disease associations.
Hum Genet
110
208
17. HegeleR
2002
SNP judgements and freedom of association.
Arterioscler Thromb Vasc Biol
22
1058
1061
18. LittleJBradleyLBrayMSClyneMDormanJ
2002
Reporting, appraising, and integrating data on genotype prevalence and gene-disease associations.
Am J Epidemiol
156
300
310
19. RomeroRKuivaniemiHTrompGOlsonJM
2002
The design, execution, and interpretation of genetic association studies to decipher complex diseases.
Am J Obstet Gynecol
187
1299
1312
20. ColhounHMMcKeiguePMDavey SmithG
2003
Problems of reporting genetic associations with complex outcomes.
Lancet
361
865
872
21. van DuijnCMPortaM
2003
Good prospects for genetic and molecular epidemiologic studies in the European Journal of Epidemiology.
Eur J Epidemiol
18
285
286
22. CrossmanDWatkinsH
2004
Jesting Pilate, genetic case-control association studies, and Heart.
Heart
90
831
832
23. HuizingaTWPisetskyDSKimberlyRP
2004
Associations, populations, and the truth: Recommendations for genetic association studies in arthritis & rheumatism.
Arthritis Rheum
50
2066
2071
24. LittleJ
2004
Reporting and review of human genome epidemiology studies.
KhouryMJLittleJBurkeW
editors In
Human genome epidemiology: A scientific foundation for using genetic information to improve health and prevent disease
New York
Oxford University Press
168
192
25. RebbeckTRMartinezMESellersTAShieldsPGWildCP
2004
Genetic variation and cancer: Improving the environment for publication of association studies.
Cancer Epidemiol Biomarkers Prev
13
1985
1986
26. TanNMulleyJBerkovicS
2004
Association studies in epilepsy: “The truth is out there”.
Epilepsia
45
1429
1442
27. [Anonymous]
2005
Framework for a fully powered risk engine.
Nat Genet
37
1153
28. EhmMGNelsonMRSpurrNK
2005
Guidelines for conducting and reporting whole genome/large-scale association studies.
Hum Mol Genet
14
2485
2488
29. FreimerNBSabattiC
2005
Guidelines for association studies in human molecular genetics.
Hum Mol Genet
14
2481
2483
30. HattersleyATMcCarthyMI
2005
What makes a good genetic association study?
Lancet
366
1315
1323
31. ManlyK
2005
Reliability of statistical associations between genes and disease.
Immunogenetics
57
549
558
32. ShenHLiuYLiuPReckerRDengH
2005
Nonreplication in genetic studies of complex diseases—Lessons learned from studies of osteoporosis and tentative remedies.
J Bone Miner Res
20
365
376
33. VitaliSRandolphA
2005
Assessing the quality of case-control association studies on the genetic basis of sepsis.
Pediatr Crit Care Med
6
S74
S77
34. WedzichaJAHallIP
2005
Publishing genetic association studies in Thorax.
Thorax
60
357
35. HallIPBlakeyJD
2005
Genetic association studies in Thorax.
Thorax
60
357
359
36. DeLisiLEFaraoneSV
2006
When is a “positive” association truly a “positive” in psychiatric genetics? A commentary based on issues debated at the world congress of psychiatric genetics, Boston, October 12–18, 2005.
Am J Med Genet B Neuropsychiatr Genet
141
319
322
37. SaitoYATalleyNJde AndradeMPetersenGM
2006
Case-control genetic association studies in gastrointestinal disease: Review and recommendations.
Am J Gastroenterol
101
1379
1389
38. UhligKMenonVSchmidCH
2007
Recommendations for reporting of clinical research studies.
Am J Kidney Dis
49
3
7
39. NCI-NHGRI Working Group on Replication in Association Studies
ChanockSJManolioTBoehnkeMBoerwinkleE
2007
Replicating genotype-phenotype associations.
Nature
447
655
660
40. BeggCB
2005
Reflections on publication criteria for genetic association studies.
Cancer Epidemiol Biomarkers Prev
14
1364
1365
41. ByrnesGGurrinLDowtyJHopperJL
2005
Publication policy or publication bias?
Cancer Epidemiol Biomarkers Prev
14
1363
42. PharoahPDDunningAMPonderBAEastonDF
2005
The reliable identification of disease-gene associations.
Cancer Epidemiol Biomarkers Prev
14
1362
43. WacholderS
2005
Publication environment and broad investigation of the genome.
Cancer Epidemiol Biomarkers Prev
14
1361
44. WhittemoreAS
2005
Genetic association studies: Time for a new paradigm?
Cancer Epidemiol Biomarkers Prev
14
1359
1360
45. BogardusSTJrConcatoJFeinsteinAR
1999
Clinical epidemiological quality in molecular genetic research. The need for methodological standards.
JAMA
281
1919
1926
46. PetersDLBarberRCFloodEMGarnerHRO'KeefeGE
2003
Methodologic quality and genotyping reproducibility in studies of tumor necrosis factor –308 G–>A single nucleotide polymorphism and bacterial sepsis: Implications for studies of complex traits.
Crit Care Med
31
1691
1696
47. ClarkMFBaudouinSV
2006
A systematic review of the quality of genetic association studies in human sepsis.
Intensive Care Med
32
1706
1712
48. LeeWBindmanJFordTGlozierNMoranP
2007
Bias in psychiatric case-control studies: Literature survey.
Br J Psychiatry
190
204
209
49. YesupriyaAEvangelouEKavvouraFKPatsopoulosNAClyneM
2008
Reporting of human genome epidemiology (HuGE) association studies: An empirical assessment.
BMC Med Res Methodol
8
31
50. ReidMCLachsMSFeinsteinAR
1995
Use of methodological standards in diagnostic test research. Getting better but still not good.
JAMA
274
645
651
51. BrazmaAHingampPQuackenbushJSherlockGSpellmanP
2001
Minimum information about a microarray experiment (MIAME)—Toward standards for microarray data.
Nat Genet
29
356
371
52. PocockSJCollierTJDandreoKJde StavolaBLGoldmanMB
2004
Issues in the reporting of epidemiological studies: A survey of recent practice.
BMJ
329
883
53. AltmanDMoherD
2005
Developing guidelines for reporting healthcare research: Scientific rationale and procedures.
Med Clin (Barc)
125
8
13
54. GluudLL
2006
Bias in clinical intervention research.
Am J Epidemiol
163
493
501
55. von ElmEAltmanDGEggerMPocockSJGotzschePC
2007
The strengthening the reporting of observational studies in epidemiology (STROBE) statement: Guidelines for reporting observational studies.
PLoS Med
4
e296
doi:10.1371/journal.pmed.0040296
56. VandenbrouckeJPvon ElmEAltmanDGGotzschePCMulrowCD
2007
Strengthening the reporting of observational studies in epidemiology (STROBE): Explanation and elaboration.
Ann Intern Med
147
W163
W194
57. LittleJHigginsJPT
editors
2006
The HuGENet™ HuGE Review Handbook, version 1.0.
Available: http://www.hugenet.ca. Accessed 28 February 2006
58. HigginsJPLittleJIoannidisJPBrayMSManolioTA
2007
Turning the pump handle: Evolving methods for integrating the evidence on gene-disease association.
Am J Epidemiol
166
863
866
59. AltmanDGSchulzKFMoherDEggerMDavidoffF
2001
The revised CONSORT statement for reporting randomized trials: Explanation and elaboration.
Ann Intern Med
134
663
694
60. MoherDSchultzKFAltmanD
2001
The CONSORT statement: Revised recommendations for improving the quality of reports of parallel-group randomized trials.
JAMA
285
1987
1991
61. PompanonFBoninABellemainETaberletP
2005
Genotyping errors: Causes, consequences and solutions.
Nat Rev Genet
6
847
859
62. AkeyJMZhangKXiongMDorisPJinL
2001
The effect that genotyping errors have on the robustness of common linkage-disequilibrium measures.
Am J Hum Genet
68
1447
1456
63. DequekerERamsdenSGrodyWWStenzelTTBartonDE
2001
Quality control in molecular genetic testing.
Nat Rev Genet
2
717
723
64. MitchellAACutlerDJChakravartiA
2003
Undetected genotyping errors cause apparent overtransmission of common alleles in the transmission/disequilibrium test.
Am J Hum Genet
72
598
610
65. RothmanNStewartWFCaporasoNEHayesRB
1993
Misclassification of genetic susceptibility biomarkers: Implications for case-control studies and cross-population comparisons.
Cancer Epidemiol Biomarkers Prev
2
299
303
66. Garcia-ClosasMWacholderSCaporasoNRothmanN
2004
Inference issues in cohort and case-control studies of genetic effects and gene-environment interactions.
KhouryMJLittleJBurkeW
editors. In
Human genome epidemiology: A scientific foundation for using genetic information to improve health and prevent disease
New York
Oxford University Press
127
144
67. WongMYDayNELuanJAWarehamNJ
2004
Estimation of magnitude in gene-environment interactions in the presence of measurement error.
Stat Med
23
987
998
68. ClaytonDGWalkerNMSmythDJPaskRCooperJD
2005
Population structure, differential bias and genomic control in a large-scale, case-control association study.
Nat Genet
37
1243
1246
69. KnowlerWCWilliamsRCPettittDJSteinbergAG
1988
Gm3;5,13,14 and type 2 diabetes mellitus: An association in American Indians with genetic admixture.
Am J Human Genet
43
520
526
70. GelernterJGoldmanDRischN
1993
The A1 allele at the D2 dopamine receptor gene and alcoholism: A reappraisal.
JAMA
269
1673
1677
71. KittlesRAChenWPanguluriRKAhaghotuCJacksonA
2002
CYP3A4-V and prostate cancer in African Americans: Causal or confounding association because of population stratification?
Hum Genet
110
553
560
72. ThomasDCWitteJS
2002
Point: Population stratification: A problem for case control studies of candidate-gene associations?
Cancer Epidemiol Biomarkers Prev
11
505
512
73. WacholderSChatterjeeNHartgeP
2002
Joint effects of genes and environment distorted by selection biases: Implications for hospital-based case-control studies.
Cancer Epidemiol Biomarkers Prev
11
885
889
74. CardonLRPalmerLJ
2003
Population stratification and spurious allelic association.
Lancet
361
598
604
75. WacholderSRothmanNCaporasoN
2000
Population stratification in epidemiologic studies of common genetic variants and cancer: Quantification of bias.
J Natl Cancer Inst
92
1151
1158
76. ArdlieKGLunettaKLSeielstadM
2002
Testing for population subdivision and association in four case-control studies.
Am J Human Genet
71
304
311
77. EdlandSDSlagerSFarrerM
2004
Genetic association studies in Alzheimer's disease research: Challenges and opportunities.
Stat Med
23
169
178
78. MillikanRC
2001
Re: Population stratification in epidemiologic studies of common genetic variants and cancer: Quantification of bias.
J Natl Cancer Inst
93
156
157
79. WangYLocalioRRebbeckTR
2004
Evaluating bias due to population stratification in case-control association studies of admixed populations.
Genet Epidemiol
27
14
20
80. IoannidisJPNtzaniEETrikalinosTA
2004
‘Racial’ differences in genetic effects for complex diseases.
Nat Genet
36
1312
1318
81. MarchiniJCardonLRPhillipsMSDonnellyP
2004
The effects of human population structure on large genetic association studies.
Nat Genet
36
512
517
82. FreedmanMLReichDPenneyKLMcDonaldGJMignaultAA
2004
Assessing the impact of population stratification on genetic association studies.
Nat Genet
36
388
393
83. KhlatMCazesMHGeninEGuiguetM
2004
Robustness of case-control studies of genetic factors to population stratification: Magnitude of bias and type I error.
Cancer Epidemiol Biomarkers Prev
13
1660
1664
84. BaldingDJ
2006
A tutorial on statistical methods for population association studies.
Nat Rev Genet
7
781
791
85. Wellcome Trust Case Control Consortium
2007
Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls.
Nature
447
661
678
86. IoannidisJP
2007
Non-replication and inconsistency in the genome-wide association setting.
Hum Hered
64
203
213
87. ParkesMBarrettJCPrescottNJTremellingMAndersonCA
2007
Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility.
Nat Genet
39
830
832
88. ToddJAWalkerNMCooperJDSmythDJDownesK
2007
Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes.
Nat Genet
39
857
864
89. ZegginiEWeedonMNLindgrenCMFraylingTMElliottKS
2007
Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes.
Science
316
1336
1341
90. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT Lund University, and Novartis Institutes of BioMedical Research
SaxenaRVoightBFLyssenkoVBurttNP
2007
Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels.
Science
316
1331
1336
91. ScottLJMohlkeKLBonnycastleLLWillerCJLiY
2007
A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants.
Science
316
1341
1345
92. HelgadottirAThorleifssonGManolescuAGretarsdottirSBlondalT
2007
A common variant on chromosome 9p21 affects the risk of myocardial infarction.
Science
316
1491
1493
93. McPhersonRPertsemlidisAKavaslarNStewartARobertsR
2007
A common allele on chromosome 9 associated with coronary heart disease.
Science
316
1488
1491
94. EastonDFPooleyKADunningAMPharoahPDThompsonD
2007
Genome-wide association study identifies novel breast cancer susceptibility loci.
Nature
447
1087
1093
95. HunterDJKraftPJacobsKBCoxDGYeagerM
2007
A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer.
Nat Genet
39
870
874
96. StaceySNManolescuASulemPRafnarTGudmundssonJ
2007
Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer.
Nat Genet
39
865
869
97. GudmundssonJSulemPSteinthorsdottirVBergthorssonJTThorleifssonG
2007
Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes.
Nat Genet
39
977
983
98. HaimanCAPattersonNFreedmanMLMyersSRPikeMC
2007
Multiple regions within 8q24 independently affect risk for prostate cancer.
Nat Genet
39
638
644
99. YeagerMOrrNHayesRBJacobsKBKraftP
2007
Genome-wide association study of prostate cancer identifies a second risk locus at 8q24.
Nat Genet
39
645
649
100. ZankeBWGreenwoodCMRangrejJKustraRTenesaA
2007
Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24.
Nat Genet
39
989
994
101. TomlinsonIWebbECarvajal-CarmonaLBroderickPKempZ
2007
A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21.
Nat Genet
39
984
988
102. HaimanCALe MarchandLYamamotoJStramDOShengX
2007
A common genetic risk factor for colorectal and prostate cancer.
Nature Genetics
39
954
956
103. RiouxJDXavierRJTaylorKDSilverbergMSGoyetteP
2007
Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis.
Nat Genet
39
596
604
104. LibioulleCLouisEHansoulSSandorCFarnirF
2007
Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4.
PLoS Genet
3
e58
doi:10.1371/journal.pgen.0030058
105. DuerrRHTaylorKDBrantSRRiouxJDSilverbergMS
2006
A genome-wide association study identifies IL23R as an inflammatory bowel disease gene.
Science
314
1461
1463
106. ZhaoLPLiSSKhalidN
2003
A method for the assessment of disease associations with single-nucleotide polymorphism haplotypes and environmental variables in case-control studies.
Am J Hum Genet
72
1231
1250
107. International HapMap Consortium
FrazerKABallingerDGCoxDRHindsDA
2007
A second generation human haplotype map of over 3.1 million SNPs.
Nature
449
851
861
108. StephensMSmithNJDonnellyP
2001
A new statistical method for haplotype reconstruction from population data.
Am J Hum Genet
68
978
989
109. QinZSNiuTLiuJS
2002
Partition-ligation-expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms.
Am J Hum Genet
71
1242
1247
110. ScheetPStephensM
2006
A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase.
Am J Hum Genet
78
629
644
111. BrowningSR
2008
Missing data imputation and haplotype phase inference for genome-wide association studies.
Hum Genet
124
439
450
112. HuangQFuYXBoerwinkleE
2003
Comparison of strategies for selecting single nucleotide polymorphisms for case/control association studies.
Hum Genet
113
253
257
113. KamataniNSekineAKitamotoTIidaASaitoS
2004
Large-scale single-nucleotide polymorphism (SNP) and haplotype analyses, using dense SNP maps, of 199 drug-related genes in 752 subjects: The analysis of the association between uncommon SNPs within haplotype blocks and the haplotypes constructed with haplotype-tagging SNPs.
Am J Hum Genet
75
190
203
114. ZhangWCollinsAMortonNE
2004
Does haplotype diversity predict power for association mapping of disease susceptibility?
Hum Genet
115
157
164
115. CarlsonCSEberleMARiederMJYiQKruglyakL
2004
Selecting a maximally informative set of single-nucleotide polymorphisms for association analysis using linkage disequilibrium.
Am J Hum Genet
74
106
120
116. van Hylckama VliegASandkuijlLARosendaalFRBertinaRMVosHL
2004
Candidate gene approach in association studies: Would the factor V leiden mutation have been found by this approach?
Eur J Hum Genet
12
478
482
117. GreenspanGGeigerD
2004
Model-based inference of haplotype block variation.
J Comput Biol
11
493
504
118. KimmelGShamirR
2005
GERBIL: Genotype resolution and block identification using likelihood.
Proc Natl Acad Sci U S A
102
158
162
119. CardonLRAbecasisGR
2003
Using haplotype blocks to map human complex triat loci.
Trends Genet
19
135
140
120. KeXHuntSTapperWLawrenceRStavridesG
2004
The impact of SNP density on fine-scale patterns of linkage disequilibrium.
Hum Mol Genet
13
577
588
121. ServinBStephensM
2007
Imputation-based analysis of association studies: Candidate regions and quantitative traits.
PLoS Genet
3
e114
doi:10.1371/journal.pgen.0030114
122. MarchiniJHowieBMyersSMcVeanGDonnellyP
2007
A new multipoint method for genome-wide association studies by imputation of genotypes.
Nat Genet
39
906
913
123. HardyGH
1908
Mendelian proportions in a mixed population.
Science
28
49
50
124. WeinbergW
1908
Über den nachweis der vererbung beim menschen.
Jahrhefte Des Vereines Für Vaterländische Naturkunde in Württemberg
64
368
382
125. MinelliCThompsonJRAbramsKRThakkinstianAAttiaJ
2008
How should we use information about HWE in the meta-analyses of genetic association studies?
Int J Epidemiol
37
136
146
126. XuJTurnerALittleJBleeckerERMeyersDA
2002
Positive results in association studies are associated with departure from Hardy-Weinberg equilibrium: Hint for genotyping error?
Hum Genet
111
573
574
127. HoskingLLumsdenSLewisKYeoAMcCarthyL
2004
Detection of genotyping errors by Hardy-Weinberg equilibrium testing.
Eur J Hum Genet
12
395
399
128. SalantiGAmountzaGNtzaniEEIoannidisJP
2005
Hardy-Weinberg equilibrium in genetic association studies: An empirical evaluation of reporting, deviations, and power.
Eur J Hum Genet
13
840
848
129. PearsonTAManolioTA
2008
How to interpret a genome-wide association study.
JAMA
299
1335
1344
130. McCarthyMIAbecasisGRCardonLRGoldsteinDBLittleJ
2008
Genome-wide association studies for complex traits: Consensus, uncertainty and challenges.
Nat Rev Genet
9
356
369
131. ZouGYDonnerA
2006
The merits of testing Hardy-Weinberg equilibrium in the analysis of unmatched case-control data: A cautionary note.
Ann Hum Genet
70
923
933
132. ShoemakerJPainterIWeirBS
1998
A Bayesian characterization of Hardy-Weinberg disequilibrium.
Genetics
149
2079
2088
133. AyresKLBaldingDJ
1998
Measuring departures from Hardy-Weinberg: A Markov chain Monte Carlo method for estimating the inbreeding coefficient.
Heredity
80
769
777
134. TrikalinosTASalantiGKhouryMJIoannidisJP
2006
Impact of violations and deviations in Hardy-Weinberg equilibrium on postulated gene-disease associations.
Am J Epidemiol
163
300
309
135. DavidoffFBataldenPStevensDOgrincGMooneyS
2008
Publication guidelines for improvement studies in health care: Evolution of the SQUIRE project.
Ann Intern Med
149
670
676
136. SteinbergKGallagherM
2004
Assessing genotypes in human genome epidemiology studies.
KhouryMJLittleJBurkeW
editors. In
Human genome epidemiology: A scientific foundation for using genetic information to improve health and prevent disease
New York
Oxford University Press
79
91
137. PlagnolVCooperJDToddJAClaytonDG
2007
A method to address differential bias in genotyping in large-scale association studies.
PLoS Genet
3
e74
doi:10.1371/journal.pgen.0030074
138. WinkerMA
2006
Race and ethnicity in medical research: Requirements meet reality.
J Law Med Ethics
34
520
525
139. ScuteriASannaSChenWMUdaMAlbaiG
2007
Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits.
PLoS Genet
3
e115
doi:10.1371/journal.pgen.0030115
140. ChanAWHrobjartssonAHaahrMTGotzschePCAltmanDG
2004
Empirical evidence for selective reporting of outcomes in randomized trials: Comparison of protocols to published articles.
JAMA
291
2457
2465
141. ChanAWKrleza-JericKSchmidIAltmanDG
2004
Outcome reporting bias in randomized trials funded by the Canadian Institutes of Health Research.
CMAJ
171
735
740
142. ChanAWAltmanDG
2005
Identifying outcome reporting bias in randomised trials on PubMed: Review of publications and survey of authors.
BMJ
330
753
143. Contopoulos-IoannidisDGAlexiouGAGouviasTCIoannidisJP
2006
An empirical evaluation of multifarious outcomes in pharmacogenetics: Beta-2 adrenoceptor gene polymorphisms in asthma treatment.
Pharmacogenet Genomics
16
705
711
144. WainHMBrufordEALoveringRCLushMJWrightMW
2002
Guidelines for human gene nomenclature.
Genomics
79
464
470
145. WainHMLushMDucluzeauFPoveyS
2002
Genew: The human gene nomenclature database.
Nucleic Acids Res
30
169
171
146. SherrySTWardMHKholodovMBakerJPhanL
2001
dbSNP: The NCBI database of genetic variation.
Nucleic Acids Res
29
308
311
147. AntonarakisSE
1998
Recommendations for a nomenclature system for human gene mutations. nomenclature working group.
Hum Mutat
11
1
3
148. den DunnenJTAntonarakisSE
2000
Mutation nomenclature extensions and suggestions to describe complex mutations: A discussion.
Hum Mutat
15
7
12
149. TobinMDSheehanNAScurrahKJBurtonPR
2005
Adjusting for treatment effects in studies of quantitative traits: Antihypertensive therapy and systolic blood pressure.
Stat Med
24
2911
2935
150. LynchMRitlandK
1999
Estimation of pairwise relatedness with molecular markers.
Genetics
152
1753
1766
151. SlagerSLSchaidDJ
2001
Evaluation of candidate genes in case-control studies: A statistical method to account for related subjects.
Am J Hum Genet
68
1457
1462
152. VoightBFPritchardJK
2005
Confounding from cryptic relatedness in case-control association studies.
PLoS Genet
1
e32
doi:10.1371/journal.pgen.0010032
153. HomerNSzelingerSRedmanMDugganDTembeW
2008
Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays.
PLoS Genet
4
e1000167
doi:10.1371/journal.pgen.1000167
154. ZerhouniEANabelEG
2008
Protecting aggregate genomic data.
Science
322
44
Štítky
Interní lékařství
Článek vyšel v časopisePLOS Medicine
Nejčtenější tento týden
2009 Číslo 2- Léčba bolesti u seniorů
- Benefity fixní kombinace tramadolu a paracetamolu v léčbě bolesti
- Berberin: přírodní hypolipidemikum se slibnými výsledky
- Flexofytol® – přírodní revoluce v boji proti osteoartróze kloubů
- Superoxidované roztoky v prevenci infekcí u dialyzovaných pacientů
-
Všechny články tohoto čísla
- What Should Be Done To Tackle Ghostwriting in the Medical Literature?
- An Unbiased Scientific Record Should Be Everyone's Agenda
- Post-Partum Psychosis: Which Women Are at Highest Risk?
- Malaria Control with Transgenic Mosquitoes
- Ovarian Cancer: A Clinical Challenge That Needs Some Basic Answers
- STrengthening the REporting of Genetic Association Studies (STREGA)— An Extension of the STROBE Statement
- How Do Courts Set Health Policy? The Case of the Colombian Constitutional Court
- A 21-Year-Old Pregnant Woman with Hypertension and Proteinuria
- PLOS Medicine
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- STrengthening the REporting of Genetic Association Studies (STREGA)— An Extension of the STROBE Statement
- How Do Courts Set Health Policy? The Case of the Colombian Constitutional Court
- A 21-Year-Old Pregnant Woman with Hypertension and Proteinuria
- Ovarian Cancer: A Clinical Challenge That Needs Some Basic Answers
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



