-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaStrengthening the Reporting of Genetic Risk Prediction Studies: The GRIPS Statement
article has not abstract
Published in the journal: . PLoS Med 8(3): e32767. doi:10.1371/journal.pmed.1000420
Category: Guidelines and Guidance
doi: https://doi.org/10.1371/journal.pmed.1000420Summary
article has not abstract
Summary Points
-
The rapid and continuing progress in gene discovery for complex diseases is fueling interest in the potential application of genetic risk models for clinical and public health practice.
-
The number of studies assessing the predictive ability is steadily increasing, but the quality and completeness of reporting varies.
-
A multidisciplinary workshop sponsored by the Human Genome Epidemiology Network developed a checklist of 25 items recommended for strengthening the reporting of Genetic RIsk Prediction Studies (GRIPS), building on the principles established by prior reporting guidelines.
-
These recommendations aim to enhance the transparency of study reporting, and thereby to improve the synthesis and application of information from multiple studies that might differ in design, conduct, or analysis.
-
A detailed Explanation and Elaboration document is published as supporting information (Text S1).
Introduction
The recent successes of genome-wide association studies and the promises of whole genome sequencing fuel interest in the translation of this new wave of basic genetic knowledge to health care practice. Knowledge about genetic risk factors may be used to target diagnostic, preventive, and therapeutic interventions for complex disorders based on a person’s genetic risk, or to complement existing risk models based on classical nongenetic factors such as the Framingham risk score for cardiovascular disease. Implementation of genetic risk prediction in health care requires a series of studies that encompass all phases of translational research [1],[2], starting with a comprehensive evaluation of genetic risk prediction.
With increasing numbers of discovered genetic markers that can be used in future genetic risk prediction studies, it is crucial to enhance the quality of the reporting of these studies, since valid interpretation could be compromised by the lack of reporting of key information. Information that is often missing includes details in the description of how the study was designed and conducted (e.g., how genetic variants were selected and coded, how risk models or genetic risk scores were constructed, and how risk categories were chosen), or how the results should be interpreted. An appropriate assessment of the study’s strengths and weaknesses is not possible without this information. There is ample evidence that prediction research often suffers from poor design and bias, and these may also have an impact on the results of the studies and on models of disease outcomes based on these studies [3]–[5]. Although most prognostic studies published to date claim significant results [6],[7], very few translate to clinically useful applications. Just as for observational epidemiological studies [8], poor reporting complicates the use of the specific study for research, clinical, or public health purposes and hampers the synthesis of evidence across studies.
Reporting guidelines have been published for various research designs [9], and these contain many items that are also relevant to genetic risk prediction studies. In particular, the guidelines for genetic association studies (STREGA) have relevant items on the assessment of genetic variants, and the guidelines for observational studies (STROBE) have relevant items about the reporting of study design. The guidelines for diagnostic studies (STARD) and those for tumor marker prognostic studies (REMARK) include relevant items about test evaluation; the REMARK guidelines also have relevant items about risk prediction [10]–[13]. However, none of these guidelines are fully suited to genetic risk prediction studies, an emerging field of investigation with specific methodological issues that need to be addressed, such as the handling of large numbers of genetic variants (from 10s to 10,000s) and flexibility in handling such large numbers in analyses. We organized a two-day workshop with an international group of risk prediction researchers, epidemiologists, geneticists, methodologists, statisticians, and journal editors to develop recommendations for the reporting of Genetic RIsk Prediction Studies (GRIPS).
Genetic Risk Prediction Studies
Genetic risk prediction studies typically develop or validate models that predict the risk of disease, but they are also being investigated for use in predicting prognostic outcome, treatment response, or treatment-related harms. Risk prediction models are statistical algorithms, which may be simple genetic risk scores (e.g., risk allele counts), may be based on regression analyses (e.g., weighted risk scores or predicted risks), or may be based on more complex analytic approaches such as support vector machine learning or classification trees. The risk models may be based on genetic variants only, or include both genetic and nongenetic risk factors [14].
Aims and Use of the GRIPS Statement
The 25 items of the GRIPS statement are intended to maximize the transparency, quality, and completeness of reporting on research methodology and findings in a particular study. It is important to emphasize that these recommendations are guidelines only for how to report research and do not prescribe how to perform genetic risk prediction studies. The guidelines do not support or oppose the choice of any particular study design or method, e.g., the guidelines recommend that the study population should be described, but do not specify which population is preferred in a particular study.
The intended audience for the reporting guidelines is broad and includes epidemiologists, geneticists, statisticians, clinician scientists, and laboratory-based investigators who undertake genetic risk prediction studies, as well as journal editors and reviewers who have to appraise the design, conduct and analysis of such studies. In addition, it includes “users” of such studies who wish to understand the basic premise, design, and limitations of genetic prediction studies in order to interpret the results for their potential application in health care. These guidelines are also intended to ensure that essential data from future genetic risk prediction studies are presented in standardized form, which will facilitate information synthesis as part of systematic reviews and meta-analyses.
Items presented in the checklist are relevant for a wide array of risk prediction studies, because GRIPS focuses on the main aspects of the design and analysis of risk prediction studies. GRIPS does not address randomized trials that may be performed to test risk models, nor does it specifically address decision analyses, cost-effectiveness analyses, assessment of health care needs, or assessment of barriers to health care implementation [15]. Once the performance of a risk model has been established, these next steps toward implementation require further evaluation [10],[16]. For the reporting of these studies, which go beyond the assessment of genetic risk models as such, additional requirements apply. However, proper documentation of genetic predictive research according to GRIPS might facilitate the translation of research findings into clinical and public health practice.
Development of the GRIPS Statement
The GRIPS statement was developed by a multidisciplinary panel of 25 risk prediction researchers, epidemiologists, geneticists, methodologists, statisticians, and journal editors, seven of whom were also part of the STREGA initiative [11]. They attended a two-day meeting in Atlanta, Georgia (US) in December 2009 that was sponsored by the US Centers for Disease Control and Prevention on behalf of the Human Genome Epidemiology Network (HuGENet) [17]. Participants discussed a draft version of the guidelines that was prepared and distributed before the meeting. This draft version was developed on the basis of existing reporting guidelines, namely STREGA [11], REMARK [13], and STARD [12]. These were selected out of all available guidelines (see http://www.equator-network.org) because of their focus on observational study designs and genetic factors (STREGA), prediction models (REMARK), and test evaluation (REMARK and STARD). During the meeting, methodological issues pertinent to risk prediction studies were addressed in presentations. Workshop participants were asked to change, combine, or delete proposed items and add additional items if necessary. Participants had extensive post-meeting electronic correspondence. To harmonize our recommendations for genetic risk prediction studies with previous guidelines, we chose the same wording for the items wherever possible. Finally, we tried to create consistency with previous guidelines for the evaluation of risk prediction studies of cardiovascular diseases and cancer [2],[18]. The final version of the checklist is presented in Table 1.
Tab. 1. Reporting recommendations for evaluations of risk prediction models that include genetic variants. 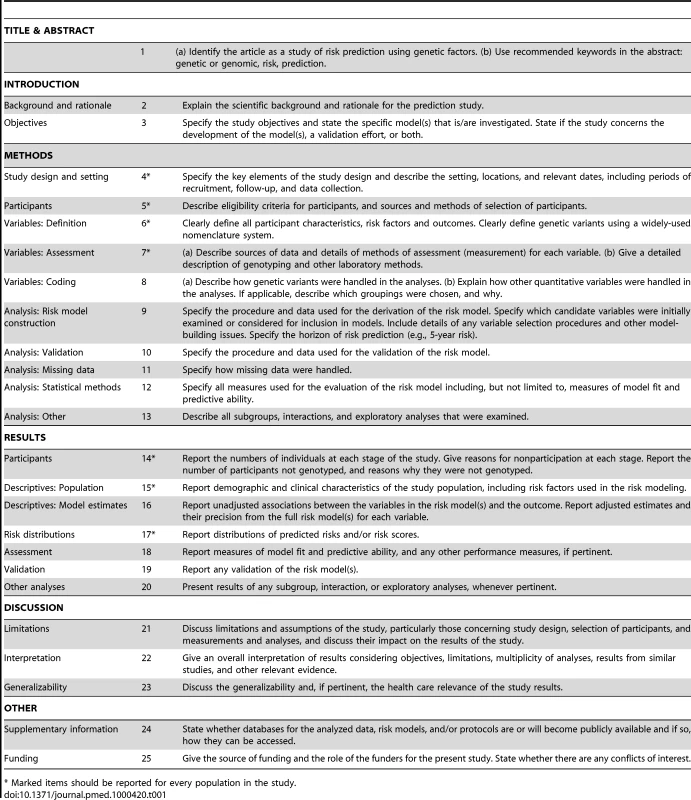
* Marked items should be reported for every population in the study. The GRIPS Explanation and Elaboration Article
Accompanying this GRIPS statement, an Explanation and Elaboration document has been written (see Text S1), modeled after those developed for other reporting guidelines [19]–[22]. The Explanation and Elaboration document illustrates each item with at least one published example that we consider transparent in reporting, explains the rationale for its inclusion in the checklist, and presents details of the items that need to be addressed to ensure transparent reporting. The Explanation and Elaboration document was produced after the meeting. The document was prepared by a small subgroup and shared with all workshop participants for additional revisions and final approval.
Concluding Remarks and Future Directions
High-quality reporting reveals the strengths and weaknesses of empirical studies, facilitates the interpretation of the scientific and health care relevance of the results—especially within the framework of systematic reviews and meta-analyses—and helps build a solid evidence base for moving genomic discoveries into applications in health care practice. The GRIPS guidelines were developed to improve the transparency, quality and completeness of the reporting of genetic risk prediction studies. As outlined in the introduction, GRIPS does not prescribe how studies should be designed, conducted, or analyzed, and therefore the guidelines should not be used to assess the quality of empirical studies [23]. The guidelines should be used only to check whether all essential items are adequately reported.
Finally, the methodology for designing and assessing genetic risk prediction models is still developing. For example, newer measures of reclassification were first introduced in 2007 [24], and several alternative reclassification measures have been proposed [25]. Which measures to apply and when to use measures of reclassification are still subject to ongoing evaluation and discussion [26]. Furthermore, alternative strategies for constructing risk models other than simple regression analyses are being explored, and these may add increased complexity to the reporting. In formulating the items of the GRIPS statement, these methodological advances were anticipated. It is for this reason that the GRIPS statement recommends how a study should be reported and not how a study should be conducted or analyzed. Therefore, methodological and analytical developments will not immediately impact the validity and relevance of the items, but the GRIPS statement will be updated when this is warranted by essential new developments in the construction and evaluation of genetic risk models.
Supporting Information
Zdroje
1. KhouryMJ
GwinnM
YoonPW
DowlingN
MooreCA
2007 The continuum of translation research in genomic medicine: how can we accelerate the appropriate integration of human genome discoveries into health care and disease prevention? Genet Med 9 665 674
2. HlatkyMA
GreenlandP
ArnettDK
BallantyneCM
CriquiMH
2009 Criteria for evaluation of novel markers of cardiovascular risk: a scientific statement from the American Heart Association. Circulation 119 2408 2416
3. KyzasPA
Denaxa-KyzaD
IoannidisJP
2007 Quality of reporting of cancer prognostic marker studies: association with reported prognostic effect. J Natl Cancer Inst 99 236 243
4. KyzasPA
LoizouKT
IoannidisJP
2005 Selective reporting biases in cancer prognostic factor studies. J Natl Cancer Inst 97 1043 1055
5. McShaneLM
AltmanDG
SauerbreiW
TaubeSE
GionM
2005 REporting recommendations for tumor MARKer prognostic studies (REMARK). Nat Clin Pract Urol 2 416 422
6. KyzasPA
Denaxa-KyzaD
IoannidisJP
2007 Almost all articles on cancer prognostic markers report statistically significant results. Eur J Cancer 43 2559 2579
7. TzoulakiI
LiberopoulosG
IoannidisJP
2009 Assessment of claims of improved prediction beyond the Framingham risk score. JAMA 302 2345 2352
8. von ElmE
EggerM
2004 The scandal of poor epidemiological research. BMJ 329 868 869
9. SimeraI
MoherD
HoeyJ
SchulzKF
AltmanDG
2010 A catalogue of reporting guidelines for health research. Eur J Clin Invest 40 35 53
10. von ElmE
AltmanDG
EggerM
PocockSJ
GotzschePC
2007 The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. PLoS Med 4 e296 doi:10.1371/journal.pmed.0040296
11. LittleJ
HigginsJP
IoannidisJP
MoherD
GagnonF
2009 STrengthening the REporting of Genetic Association Studies (STREGA): an extension of the STROBE statement. PLoS Med 6 e22 doi:10.1371/journal.pmed.1000022
12. BossuytPM
ReitsmaJB
BrunsDE
GatsonisCA
GlasziouPP
2003 Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. BMJ 326 41 44
13. McShaneLM
AltmanDG
SauerbreiW
TaubeSE
GionM
2005 Reporting recommendations for tumor marker prognostic studies. J Clin Oncol 23 9067 9072
14. JanssensACJW
Van DuijnCM
2009 Genome-based prediction of common diseases: methodological considerations for future research. Genome Med 1 20
15. KhouryMJ
GwinnM
IoannidisJP
2010 The emergence of translational epidemiology: from scientific discovery to population health impact. Am J Epidemiol 172 517 524
16. MoonsKG
AltmanDG
VergouweY
RoystonP
2009 Prognosis and prognostic research: application and impact of prognostic models in clinical practice. BMJ 338 b606
17. KhouryMJ
DormanJS
1998 The Human Genome Epidemiology Network. Am J Epidemiol 148 1 3
18. FreedmanAN
SeminaraD
GailMH
HartgeP
ColditzGA
2005 Cancer risk prediction models: a workshop on development, evaluation, and application. J Natl Cancer Inst 97 715 723
19. AltmanDG
SchulzKF
MoherD
EggerM
DavidoffF
2001 The revised CONSORT statement for reporting randomized trials: explanation and elaboration. Ann Intern Med 134 663 694
20. BossuytPM
ReitsmaJB
BrunsDE
GatsonisCA
GlasziouPP
2003 The STARD statement for reporting studies of diagnostic accuracy: explanation and elaboration. Ann Intern Med 138 W1 12
21. LiberatiA
AltmanDG
TetzlaffJ
MulrowC
GotzschePC
2009 The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med 6 e1000100 doi:10.1371/journal.pmed.1000100
22. VandenbrouckeJP
von ElmE
AltmanDG
GotzschePC
MulrowCD
2007 Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): explanation and elaboration. PLoS Med 4 e297 doi:10.1371/journal.pmed.0040297
23. VandenbrouckeJP
2009 STREGA, STROBE, STARD, SQUIRE, MOOSE, PRISMA, GNOSIS, TREND, ORION, COREQ, QUOROM, REMARK... and CONSORT: for whom does the guideline toll? J Clin Epidemiol 62 594 596
24. CookNR
2007 Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation 115 928 935
25. PencinaMJ
D'AgostinoRBSr
D'AgostinoRBJr
VasanRS
2008 Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 27 157 172
26. JanssensACJW
KhouryMJ
2010 Assessment of improved prediction beyond traditional risk factors: when does a difference make a difference? Circ: Cardiovasc Genet 3 3 5
Štítky
Interní lékařství
Článek vyšel v časopisePLOS Medicine
Nejčtenější tento týden
2011 Číslo 3- Příznivý vliv Armolipidu Plus na hladinu cholesterolu a zánětlivé parametry u pacientů s chronickým subklinickým zánětem
- Berberin: přírodní hypolipidemikum se slibnými výsledky
- Léčba bolesti u seniorů
- Nutraceutikum Armolipid Plus podle klinických důkazů zlepšuje lipidový profil − metaanalýza
- Snižuje terapie betablokátory kardiovaskulární benefit aerobního cvičení u pacientů s arteriální hypertenzí?
-
Všechny články tohoto čísla
- Strengthening the Reporting of Genetic Risk Prediction Studies: The GRIPS Statement
- Towards Open and Equitable Access to Research and Knowledge for Development
- HIV-1 Drug Resistance Emergence among Breastfeeding Infants Born to HIV-Infected Mothers during a Single-Arm Trial of Triple-Antiretroviral Prophylaxis for Prevention of Mother-To-Child Transmission: A Secondary Analysis
- The BCG World Atlas: A Database of Global BCG Vaccination Policies and Practices
- Promotional Tone in Reviews of Menopausal Hormone Therapy After the Women's Health Initiative: An Analysis of Published Articles
- How Can Institutional Review Boards Best Interpret Preclinical Data?
- Equity Must Accompany Economic Growth for Good Health
- Predicting Harms and Benefits in Translational Trials: Ethics, Evidence, and Uncertainty
- Effectiveness of the Standard WHO Recommended Retreatment Regimen (Category II) for Tuberculosis in Kampala, Uganda: A Prospective Cohort Study
- Scaling Up Diarrhea Prevention and Treatment Interventions: A Lives Saved Tool Analysis
- Triple-Antiretroviral Prophylaxis to Prevent Mother-To-Child HIV Transmission through Breastfeeding—The Kisumu Breastfeeding Study, Kenya: A Clinical Trial
- Is Economic Growth Associated with Reduction in Child Undernutrition in India?
- Mutations in Complement Regulatory Proteins Predispose to Preeclampsia: A Genetic Analysis of the PROMISSE Cohort
- A Randomized Controlled Trial Comparing the Effects of Counseling and Alarm Device on HAART Adherence and Virologic Outcomes
- Effectiveness and Cost Effectiveness of Expanding Harm Reduction and Antiretroviral Therapy in a Mixed HIV Epidemic: A Modeling Analysis for Ukraine
- On the Path to Global Open Access: A Few More Miles to Go
- The Challenge of Discharging Research Ethics Duties in Resource-Constrained Settings
- PLOS Medicine
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The BCG World Atlas: A Database of Global BCG Vaccination Policies and Practices
- How Can Institutional Review Boards Best Interpret Preclinical Data?
- The Challenge of Discharging Research Ethics Duties in Resource-Constrained Settings
- HIV-1 Drug Resistance Emergence among Breastfeeding Infants Born to HIV-Infected Mothers during a Single-Arm Trial of Triple-Antiretroviral Prophylaxis for Prevention of Mother-To-Child Transmission: A Secondary Analysis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



