-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaViral Inhibition of the Transporter Associated with Antigen Processing (TAP): A Striking Example of Functional Convergent Evolution
Herpesviruses are large DNA viruses that are highly abundant within their host populations. Even in the presence of a healthy immune system, these viruses manage to cause lifelong infections. This persistence is partially mediated by the virus entering latency, a phase of infection characterized by limited viral protein expression. Moreover, herpesviruses have devoted a significant part of their coding capacity to immune evasion strategies. It is believed that the close coexistence of herpesviruses and their hosts has resulted in the evolution of viral proteins that specifically attack multiple arms of the host immune system. Cytotoxic T lymphocytes (CTLs) play an important role in antiviral immunity. CTLs recognize their target through viral peptides presented in the context of MHC molecules at the cell surface. Every herpesvirus studied to date encodes multiple immune evasion molecules that effectively interfere with specific steps of the MHC class I antigen presentation pathway. The transporter associated with antigen processing (TAP) plays a key role in the loading of viral peptides onto MHC class I molecules. This is reflected by the numerous ways herpesviruses have developed to block TAP function. In this review, we describe the characteristics and mechanisms of action of all known virus-encoded TAP inhibitors. Orthologs of these proteins encoded by related viruses are identified, and the conservation of TAP inhibition is discussed. A phylogenetic analysis of members of the family Herpesviridae is included to study the origin of these molecules. In addition, we discuss the characteristics of the first TAP inhibitor identified outside the herpesvirus family, namely, in cowpox virus. The strategies of TAP inhibition employed by viruses are very distinct and are likely to have been acquired independently during evolution. These findings and the recent discovery of a non-herpesvirus TAP inhibitor represent a striking example of functional convergent evolution.
Published in the journal: . PLoS Pathog 11(4): e32767. doi:10.1371/journal.ppat.1004743
Category: Review
doi: https://doi.org/10.1371/journal.ppat.1004743Summary
Herpesviruses are large DNA viruses that are highly abundant within their host populations. Even in the presence of a healthy immune system, these viruses manage to cause lifelong infections. This persistence is partially mediated by the virus entering latency, a phase of infection characterized by limited viral protein expression. Moreover, herpesviruses have devoted a significant part of their coding capacity to immune evasion strategies. It is believed that the close coexistence of herpesviruses and their hosts has resulted in the evolution of viral proteins that specifically attack multiple arms of the host immune system. Cytotoxic T lymphocytes (CTLs) play an important role in antiviral immunity. CTLs recognize their target through viral peptides presented in the context of MHC molecules at the cell surface. Every herpesvirus studied to date encodes multiple immune evasion molecules that effectively interfere with specific steps of the MHC class I antigen presentation pathway. The transporter associated with antigen processing (TAP) plays a key role in the loading of viral peptides onto MHC class I molecules. This is reflected by the numerous ways herpesviruses have developed to block TAP function. In this review, we describe the characteristics and mechanisms of action of all known virus-encoded TAP inhibitors. Orthologs of these proteins encoded by related viruses are identified, and the conservation of TAP inhibition is discussed. A phylogenetic analysis of members of the family Herpesviridae is included to study the origin of these molecules. In addition, we discuss the characteristics of the first TAP inhibitor identified outside the herpesvirus family, namely, in cowpox virus. The strategies of TAP inhibition employed by viruses are very distinct and are likely to have been acquired independently during evolution. These findings and the recent discovery of a non-herpesvirus TAP inhibitor represent a striking example of functional convergent evolution.
Introduction
The family Herpesviridae emerged approximately 400 million years ago [1]. The first members of the class Mammalia arose 200 million years ago, at around the time of the Early Jurassic period, and, since then, herpesviruses and mammals have coevolved and adapted to one another over very long periods of time. Today, members of the family Herpesviridae are numerous and widespread among not only mammals, but also many bird and reptile species; each virus displays a remarkable degree of host specificity. The longstanding interactions between virus and host have likely contributed to the development of the host’s innate and adaptive immune system and the mechanisms that viruses use to evade those systems.
The first line of defense against intruding pathogens is the innate immune system. This comprises the complement system, natural killer (NK) cells, apoptosis, pattern recognition receptor-mediated intracellular signaling leading to the production of IFNβ and many other cytokines and chemokines, and phagocytes like neutrophils, macrophages and dendritic cells. Together, these mechanisms enable the host to limit replication and spread of a pathogen and facilitate the induction of specific adaptive immune responses. The adaptive immune system includes antibody-producing B-cells, CD4+ T-cells that recognize antigens presented in the context of MHC (class) II molecules, and CD8+ T-cells that generally recognize antigens in the context of MHC I molecules.
During and following protein synthesis, a proportion of the resulting proteins is rapidly degraded into peptides by the proteasome. The resulting peptides are subsequently translocated into the lumen of the endoplasmic reticulum (ER) via the transporter associated with antigen processing (TAP) [2,3]. Within the ER, the peptides are loaded onto newly synthesized MHC I heavy chain / β2microglobulin (β2m) heterodimers. This process is facilitated by at least five ER-resident molecules that together form the MHC I peptide-loading complex (PLC). Tapasin functions as a chaperone, bridging MHC I molecules and TAP and catalyzing the binding of high-affinity peptides [4–10]. The lectin-like chaperones calnexin and calreticulin promote folding of newly synthesized MHC I molecules; additionally, calreticulin recruits the thioloxidoreductase ERp57. ERp57 and protein disulfide isomerase (PDI) are involved in stabilizing several protein-protein interactions within the PLC via disulfide bond formation [11,12]. Acquisition of peptide allows mature MHC I complexes to leave the ER, pass through the Golgi, and traffic to the cell surface where the peptides are presented to CTLs.
The family Herpesviridae is divided into the subfamilies Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae. Members of this family have been identified in many different species, including reptiles, birds, and mammals. There are nine herpesviruses known to infect humans: herpes simplex virus (HSV) types 1 and 2 (HSV-1 and HSV-2 in species Human herpesvirus 1 and Human herpesvirus 2, respectively, of genus Simplexvirus, subfamily Alphaherpesvirinae), varicella-zoster virus (VZV in species Human herpesvirus 3 of genus Varicellovirus, subfamily Alphaherpesvirinae), human cytomegalovirus (HCMV in species Human herpesvirus 5 of genus Cytomegalovirus, subfamily Betaherpesvirinae), human herpesviruses 6A, 6B and 7 (HHV-6A, HHV-6B, HHV-7 in species Human herpesvirus 6A, Human herpesvirus 6B and Human herpesvirus 7 of genus Roseolovirus, subfamily Betaherpesvirinae), Epstein-Barr virus (EBV in species Human herpesvirus 4 of genus Lymphocryptovirus, subfamily Gammaherpesvirinae) and Kaposi’s sarcoma-associated herpesvirus (KSHV in species Human herpesvirus 8 of genus Rhadinovirus, subfamily Gammaherpesvirinae) [13]. Most of these viruses are widespread within the human population; for example, in the United States, approximately 90% of individuals of 80 years or older are seropositive for HCMV [14]. VZV is even more abundant, with a seroprevalence of 95% in people from 20 years of age [15]. Herpesvirus infections generally cause only mild symptoms, but in some circumstances they exhibit significant pathogenic properties, with the most serious complications tending to arise in immunocompromised people. Thus, HSV-1 can cause encephalitis, EBV and KSHV are associated with malignancies, and HCMV infection can result in congenital defects [16–18].
Following primary infection of their host, herpesviruses establish a state of latency in which viral protein expression is limited. As a consequence of this strategy, the virus-derived pool of potential antigens is minimized, thus hindering recognition and elimination of infected cells by CTLs and enabling the virus to persist for the lifetime of the host. In addition, several latency-associated proteins have been found actively to impede detection of virus-infected cells by the host immune system [19–21]. However, at some point, in order to disseminate the virus to other hosts, productive infection must occur. During this replicative or lytic phase, an extensive repertoire of herpesvirus-encoded genes is expressed in a kinetically regulated fashion. Depending on the virus in question, this results in the synthesis of at least 70 functional proteins, and renders the virus-infected cell susceptible to the memory immune responses that were generated during primary infection. Although these responses are instrumental in controlling infection and in limiting pathology, herpesviruses employ multiple evasion strategies to allow virus production in the face of existing antiviral immunity, thereby promoting spread within the host population.
In the past two decades, numerous articles have been published identifying and characterizing the immune evasion molecules expressed by herpesviruses. Most of these publications are focused on human herpesviruses, each of which has been shown to employ several strategies to interfere with the innate and adaptive immune responses. The MHC I presentation pathway appears to be a favorite target among the herpesviruses, illustrating the importance of CTLs in the elimination of virus-infected cells. Every step of this pathway is targeted by at least one herpesvirus. The availability of MHC I is affected by so-called host shutoff proteins that block cellular protein synthesis and thus expression of newly synthesized MHC I molecules. Examples of these proteins are the HSV-1-encoded vhs or UL41, EBV BGLF5, and KSHV SOX or ORF37 [22–26]. Additional strategies focus on inducing the degradation of MHC I molecules, as mediated by HCMV US2, US10, and US11 [27–29], murine CMV (MCMV) glycoprotein (gp) 48 [30], and murine gammaherpesvirus 68 mK3 [31–33]. Presentation of antigenic peptides by MHC I is also affected by HCMV US3 and MCMV gp40, which cause the retention of immature molecules in the cis-Golgi [34,35]. Finally, EBV BILF1 and KSHV K3 and KSHV K5 enhance the endocytosis of MHC I complexes at the cell surface [36–39].
In addition to strategies that limit the availability of MHC I, many herpesviruses affect peptide presentation by inhibiting the function of TAP. TAP is a heterodimeric ATP-binding cassette (ABC) transporter complex composed of two subunits, TAP1 and TAP2 (Fig 1). Both subunits are composed of an N-terminal transmembrane domain (TMD) and a C-terminal nucleotide-binding domain (NBD) exposed in the cytosol (Fig 1). The TMDs of TAP1 and TAP2 contain 10 and 9 transmembrane (TM) helices, respectively. The N-terminal 4 TM helices of TAP1 and the N-terminal 3 TM helices TAP2, known as TMD0s, act as autonomous interaction platforms for tapasin [40]. Together with the NBDs, the C-terminal 6 membrane helices of each TAP subunit form the core of the transporter. Expression of this core region is necessary and sufficient for peptide transport [41].
Fig. 1. Interactions between herpesvirus-encoded TAP-inhibitors and their target. 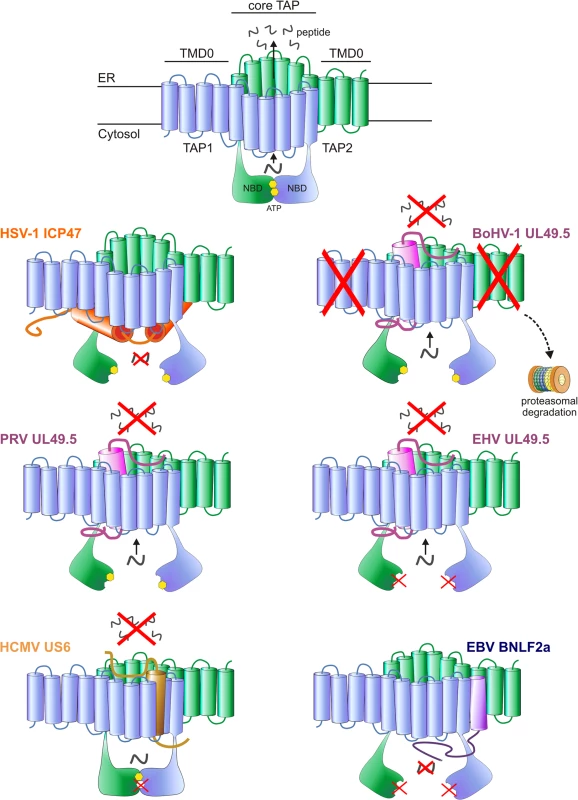
Upper illustration: model of the TAP transporter, comprising the two subunits TAP1 and TAP2. Each subunit contains a transmembrane domain (TMD), encompassing 10 and 9 transmembrane (TM) helices for TAP1 and TAP2, respectively. The outer N-terminal helices of TAP1 and TAP2 (TMD0) form an autonomous binding platform for tapasin, whereas the core 6 TM helices are necessary for peptide transport. A peptide-binding domain is located within the cytosolic extensions of the TM helices. In addition, TAP1 and TAP2 contain a nucleotide-binding domain (NBD) in the cytosol, which harbors two ATP-binding sites. Lower illustrations: schematic representations of the interaction between the viral proteins and TAP. The sites where TAP is affected are indicated. HSV-1 ICP47 prevents peptide transport by physically obstructing the peptide-binding site. PRV, BoHV-1 and EHV (EHV-1 and EHV-4) UL49.5 leave the transporter in a transformation-incompetent conformation, thereby preventing the structural changes that are needed to translocate peptides over the ER membrane. BoHV-1 UL49.5 is known to interact with a region within the core domain of TAP, comprising the C-terminal 6 TM domains of both TAP1 and TAP2 [132]. BoHV-1 UL49.5 induces the degradation of both TAP subunits, and EHV UL49.5 prevents ATP binding to TAP. HCMV US6 blocks TAP by inducing conformational changes that result in diminished ATP binding to TAP1. The protein interacts with TM domains 7–10 of TAP 1 and TM 1–4 of TAP2 [95]. EBV BNLF2a inhibits peptide transport by interfering with both peptide and ATP binding to TAP. TAP preferentially transports peptides of 8–16 amino acid residues in length, but can accommodate peptides as large as 40 amino acid residues, albeit with lower efficiency [42–46]. The exact location of the peptide-binding pocket remains unclear, but cross-linking peptide substrates to TAP and mutagenesis of TAP have shown that elements within the cytosolic extensions between the TM helices of both TAP1 and TAP2 are involved in peptide binding to TAP [47,48]. These findings are further supported by homology modeling of TAP based on the resolved crystal structures of other ABC transporters [49]. Three cytosolic pockets formed by TM helices of the TAP core complex have been proposed to represent the binding site for the peptide substrates [49].
TAP-mediated peptide transport is energized by ATP hydrolysis at the NBDs, which harbor two functionally nonequivalent ATP-binding sites. These sites are composed of conserved domains within both subunits, including the Walker A and B domains of one NBD and a signature motif of the opposing NBD. The consensus ATP-binding site that includes the Walker A and B domains of TAP2 has catalytic amino acid residues conserved among ABC transporters. The degenerate ATP-binding site, which includes the Walker A and B domains of TAP1, has a number of noncanonical mutations that reduce its catalytic activity [50]. Although ATP binding and hydrolysis can still occur at the degenerate site [51,52] only ATP binding and hydrolysis at the consensus TAP2 site is essential for completion of the transport cycle [50,52–54].
The crystal structures of several ABC transporters, trapped in distinct conformations, have been resolved [55–59]. These structures, together with biochemical studies on TAP itself, suggest that TAP transport occurs in sequential steps with extensive conformational rearrangements of the NBDs and TMDs, which depend on nucleotide and peptide substrate binding [60,61] (reviewed by [62]). In an inward-facing conformation, the peptide-binding pocket faces the cytosol and the NBDs are separated. At this stage, TAP is receptive to both peptide and ATP. The binding of peptide and ATP can occur independently and induces conformational rearrangements that partially close the NBDs. The NBDs are only fully closed when both peptide and ATP are bound. These conformational changes are relayed to the TMDs and result in an outward-facing conformation of TAP, thereby exposing the peptide-binding pocket into the ER lumen and allowing release of the peptide. Upon ATP hydrolysis, the NBDs dissociate and the TMDs rotate back into an inward-facing conformation [62]. In this way, conformational changes driven by peptide binding, ADP/ATP exchange, and ATP hydrolysis at the TAP subunits lead to the transport of peptides into the ER lumen.
Cells that naturally or experimentally lack expression of functional TAP complexes show a dramatic reduction in MHC I levels at their surface and a substantial decline in CTL sensitivity [63–68]. Herpesviruses appear to have taken advantage of this extensive dependency of MHC I expression on TAP function by encoding viral proteins that specifically impair TAP-mediated peptide transport. This review focuses on the characteristics and evolution of herpesvirus-encoded TAP inhibitors and their orthologs.
Simplexvirus ICP47 Orthologs
The first viral protein found to inhibit TAP function was HSV-1 ICP47 (Fig 1). This protein acts as a competitor of cytosolic peptides for TAP binding, thereby limiting the availability of peptides in the ER lumen and causing subsequent retention of MHC I molecules in the ER [69–72]. ICP47 is expressed as a soluble, cytosolic protein of 88 amino acid residues. Mutational analyses have defined residues 3–34 as the functional domain responsible for TAP binding and inhibition of TAP function [73–75]. Charged residues within this domain are crucial for TAP inhibition, and might mimic the N - and C-termini of peptide substrates within the peptide-binding pockets of TAP [75]. Once bound to TAP, the viral protein traps TAP in a conformation that differs from that of TAP in a peptide-bound state [76]. In contrast to peptide substrates, ICP47 blocks ATP hydrolysis at the NBDs of TAP, which is normally followed by peptide binding, suggesting that TAP is locked in an inward-facing conformation [51,77].
Orthologs of HSV-1 ICP47 are encoded by HSV-2 and other simplexviruses infecting Old World primates, including herpesvirus papio 2 (HVP-2 in species Papiine herpesvirus 2) for baboons, simian B virus (SBV in species Macacine herpesvirus 1) for macaques, and simian agent 8 (SA8 in species Cercopithecine herpesvirus 2) for African green monkeys [78]. Inhibition of TAP is conserved for HSV-2 ICP47, despite the relatively low amino acid sequence identity (44%) (Fig 2A) [79]. The ICP47 orthologs can be divided into two groups on the basis of sequence similarities within the domain responsible for TAP inhibition, i.e., amino acid residues 3 to 34 [73–75]. HSV-1 and HSV-2 ICP47 are in the first group and share identity within this domain, and both are known to inhibit TAP (Fig 2A) [79]. HPV-2, SBV, and SA8 ICP47 are in the second group. The sequences of the N-terminal domain of these proteins show substantial identity to each other and differ substantially from those of HSV-1 and HSV-2 (Fig 2A). SBV-infected cells were shown to display minor MHC I downregulation compared to HSV-1-infected cells [80], suggesting that the ICP47 protein encoded by SBV does not inhibit TAP. Given the similarities between the SBV, HPV-2, and SA8 ICP47 proteins, the HPV-2 and SA8 ICP47 proteins may also be unable to inhibit TAP, but data are not available. Thus, TAP inhibition may not be conserved for all ICP47 proteins. The fact that ICP47 is only encoded by simplexviruses suggests that this gene evolved after this lineage separated from the varicelloviruses (Fig 3). There is evidence supporting the view that the ICP47 gene arose de novo, rather than being captured from elsewhere, as is the case for many immune modulatory genes in herpesviruses [81].
Fig. 2. Alignments of the amino acid sequences of selected herpesvirus-encoded TAP-inhibitors. 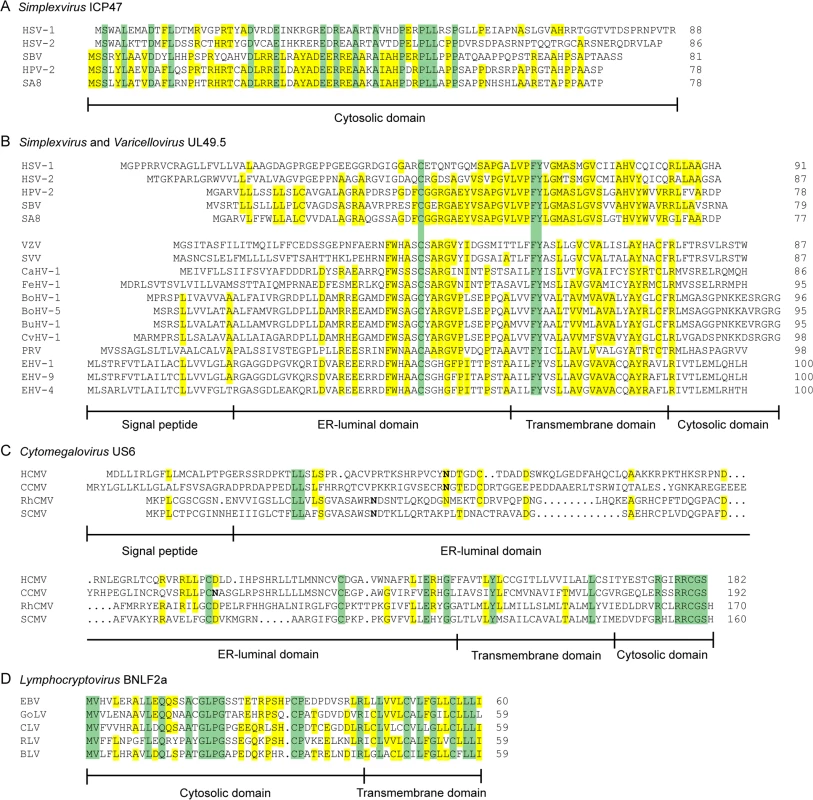
A) simplexvirus ICP47 orthologs, B) simplexvirus (upper 5 lines) and varicellovirus (lower 12 lines) UL49.5 orthologs, C) cytomegalovirus US6 orthologs, and D) lymphocryptovirus BNLF2a orthologs. The alignments of predicted primary translation products were made using ClustalW, followed by manual adjustment. The number of residues in each sequence is shown on the right. Green highlights residues that are conserved in all sequences, and yellow highlights residues that are conserved in a majority. Bold N residues in US6 indicate potential N-linked glycosylation sites. An illustrationof sequence disposition is shown below each alignment, with approximate boundaries displayed. Fig. 3. Phylogenetic tree for selected members of the family Herpesviridae. 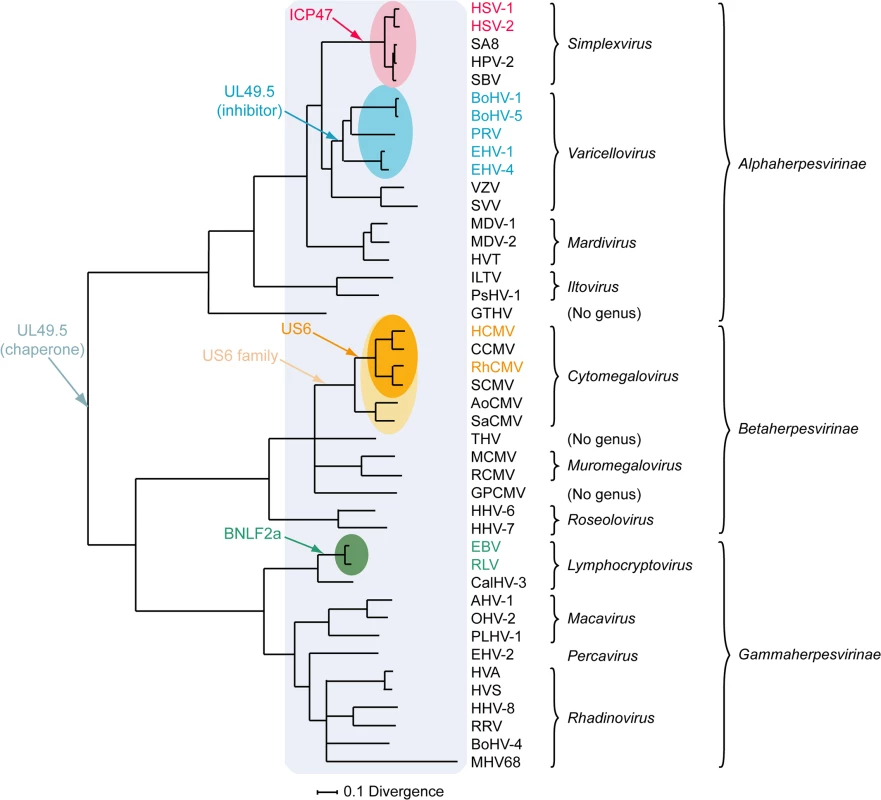
The Bayesian tree is based on amino acid sequence alignments for six large, well-conserved genes, namely the orthologs of HSV-1 genes UL15, UL19, UL27, UL28, UL29, and UL30, and is derived from McGeoch and Davison [81]. Assignments to genera and subfamilies are shown on the right. Abbreviations not mentioned in the text are: MDV-1, Marek's disease virus type 1; MDV-2, Marek's disease virus type 2; HVT, herpesvirus of turkey; ILTV, infectious laryngotracheitis virus; PsHV-1, psitticid herpesvirus 1; GTHV, green turtle herpesvirus; THV, tupaia herpesvirus; GPCMV, guinea pig cytomegalovirus; CalHV-3, callitrichine herpesvirus 3; AHV-1, alcelaphine herpesvirus 1; OHV-2, ovine herpesvirus 2; PLHV-1, porcine lymphotropic herpesvirus 1; HVS, herpesvirus saimiri; HVA, herpesvirus ateles; and RRV, rhesus rhadinovirus. Red, blue, orange, and green shading indicate viruses that encode the ICP47, UL49.5, US6, or BNLF2a TAP inhibitor genes, respectively, and corresponding coloring of virus abbreviations indicate viruses in which these genes have been shown to be functional TAP inhibitors. Light orange shading identifies all members of the Cytomegalovirus genus that have a US6 gene. Light blue shading indicates all members of the herpesvirus family that code for a UL49.5 gene that might be involved in chaperoning maturation of glycoprotein M. Varicellovirus UL49.5 Orthologs
The second TAP inhibitor discovered within the subfamily Alphaherpesvirinae is the varicellovirus UL49.5 protein. The most extensively studied member is bovine herpesvirus 1 (BoHV-1) UL49.5. Unlike ICP47, UL49.5 does not affect the binding of peptides to TAP. Instead, the viral protein binds to the core region of TAP and impairs TAP-mediated peptide transport through two unique mechanisms. First, BoHV-1 UL49.5 inhibits the conformational rearrangements that usually follow peptide and ATP binding, as inferred from fluorescence recovery after photo bleaching (FRAP) assays [82]. Using this assay, the lateral mobility of Green Fluorescent Protein (GFP)-tagged TAP can be measured in the ER-membrane. TAP mobility is affected by the conformational rearrangements that occur upon peptide transport. Upon active peptide transport, the mobility of TAP molecules is slower than that of inactive TAP [2]. In the presence of UL49.5, these changes in lateral mobility of TAP, and thus conformational rearrangements, are halted [82]. Second, BoHV-1 UL49.5 strongly reduces TAP1 and TAP2 protein levels by targeting both TAP subunits for proteasomal degradation (Fig 1) [82]. Mutational analysis of BoHV-1 UL49.5 has attributed degradation of TAP to the first five N-terminal amino acid residues and the ultimate residues of the C-terminal cytosolic domain of the protein [83]. Interestingly, UL49.5 without a C-terminal domain does not cause degradation of TAP, but retains the ability to inhibit peptide transport; this implies that the regions responsible for this effect are located in the transmembrane or ER-luminal parts of the inhibitor [82].
Orthologs of UL49.5, also known as glycoprotein N (gN), are present in all members of the family Herpesviridae sequenced to date. However, TAP inhibition by this protein has been found only among the varicelloviruses. The UL49.5 orthologs encoded by the varicelloviruses BoHV-5, bubaline herpesvirus 1 (BuHV-1), cervid herpesvirus 1 (CvHV-1), equid herpesvirus 1 (EHV-1), EHV-4, pseudorabies virus (PRV), and felid herpesvirus 1 (FeHV-1) possess the same functional properties as BoHV-1 UL49.5, causing a robust inhibition of peptide transport, thereby decreasing MHC I molecules at the cell surface [84,85]. Infections with UL49.5-deletion mutants of BoHV-1, EHV-1, and PRV have shown that UL49.5 is necessary and sufficient for TAP inhibition during viral infection in vitro [84]. Surprisingly, UL49.5 expressed by the varicelloviruses VZV, simian varicella virus (SVV), and canid herpesvirus 1 (CaHV-1) are incapable of reducing TAP function [85]. Thus, the capacity to interfere with peptide transport via TAP is a feature shared by a subgroup of varicellovirus-encoded UL49.5 orthologs.
The TAP-inhibiting UL49.5 homologs were shown to block human TAP as well as their natural host TAPs, indicating that the proteins target a highly conserved region within the TAP complex [84,85]. Each UL49.5 ortholog appears to utilize a distinct mechanism of TAP inhibition. The capacity to arrest the TAP complex in a translocation incompetent state is conserved for EHV-1 and PRV UL49.5 (Fig 1) [84] and most likely for EHV-4, BoHV-5, BuHV-1, CvHV-1, and FeHV-1 UL49.5 [85]. The EHV-1 and EHV-4 UL49.5 proteins were shown to interfere uniquely with ATP binding to TAP (Fig 1) [84]. UL49.5-induced degradation of TAP1 and TAP2 is conserved for the highly related viruses BoHV-1, BoHV-5, BuHV-1, and CvHV-1, all of which infect ruminants, but not for EHV-1, EHV-4, PRV, and FeHV-1 UL49.5 (Fig 1) [85]. The UL49.5 proteins interfering with TAP function show around 40% sequence identity (Fig 2B). Proteasomal degradation of the transporter is only induced by the ruminant-infecting viruses. The cytoplasmic domain of the TAP-degrading UL49.5 proteins contains two unique, consecutive lysine residues and an RGRG motif (Fig 2B). These lysine residues, although potential targets for ubiquitination, are not required for degradation of TAP [83]. However, the arginine residues of the RGRG sequence appeared to be essential for this phenotype [83].
Based on current knowledge, the UL49.5 molecules are the only herpesvirus-encoded TAP inhibitors that fulfill a dual role in viral infection. Within the infected cell, UL49.5 forms a heterodimeric complex with glycoprotein M (gM) and guides proper glycosylation and maturation of this protein [86–88]. Cells expressing both BoHV-1 UL49.5 and gM show reduced TAP inhibition when compared to cells expressing UL49.5 only, suggesting that the interaction between UL49.5 and gM interferes with the capacity of UL49.5 to block TAP [89]. However, UL49.5 and gM display differential temporal expression in the context of viral infection, with the appearance of UL49.5 preceding that of the late protein gM [89]. This provides UL49.5 with an opportunity to exert its immune evasive effect early during infection. Conservation of both UL49.5 and gM in the family Herpesviridae indicates that the original role of UL49.5 was that of a gM chaperone, and that TAP inhibition by the protein evolved later within the varicellovirus subfamily, possibly in the BoHV-1 lineage after its divergence from the VZV lineage (Fig 3). Alternatively, it is possible that the TAP inhibitory function was gained somewhat earlier among the alphaherpesviruses, and lost in the VZV lineage, as VZV UL49.5 is capable of interacting with the TAP complex even though it does not inhibit its activity.
Cytomegalovirus US6 Orthologs
HCMV encodes multiple immunoevasive proteins targeting the MHC I antigen presentation pathway, including the TAP inhibitor US6 [90–92]. This protein impairs TAP function by interfering with ATP binding to the transporter (Fig 1). US6 specifically prevents ATP binding to TAP1, but stimulates ATP binding to TAP2 [93]. This is in contrast with the normal pattern of ATP binding, which preferentially involves TAP1 [94]. US6 alters ATP binding to TAP by inducing conformational rearrangements that are suggested to resemble TAP in an outward-facing conformation [77,95]. Rather than physically obstructing the ATP-binding site in the cytosol, US6 induces these rearrangements by interacting with the ER luminal loops of TAP1 and TAP2 [95]. This observation is supported by data obtained with US6 truncation mutants, which shows that the ER luminal domain of US6 is necessary and sufficient for the inhibition of TAP [90,93].
Orthologs of US6 are only encoded by cytomegaloviruses infecting primates [96]. The rhesus CMV (RhCMV) ortholog of US6 (Rh185) shares only about 23% amino acid identity but nevertheless reduces cell surface expression of MHC I molecules via TAP inhibition (Fig 2C). The ER-luminal domain, identified as the functional part of US6 [90,93], shows remarkably low identity between the two orthologs (Fig 2C). HCMV US6 is a member of the US6 gene family, which is presumed to have arisen through gene duplication of a captured gene. The original gene gave rise to a block of six contiguous paralogs (US6, US7, US8, US9, US10, and US11), all encoding loosely related type I membrane proteins. Both the number of genes in this family and their encoded sequences have diverged extensively among the primate cytomegaloviruses. For example, chimpanzee cytomegalovirus (CCMV) and HCMV both encode six homologous genes in the US6 family. In contrast, RhCMV and simian CMV (SCMV) each have five genes, with orthology to HCMV being more difficult to determine [97]. Owl monkey cytomegalovirus (AoCMV) and squirrel monkey cytomegalovirus (SaCMV), which infect New World primates, have four and seven members of the US6 family, respectively. However, these are located in noncontiguous regions of the genome, and both viruses lack an obvious US6 ortholog. No US6 family genes are apparent in cytomegaloviruses of non-primate hosts, including MCMV and rat CMV (RCMV) [96]. Thus, it seems that the US6 gene family probably evolved during early primate evolution, with the TAP-inhibiting function arising in the Old World primate lineage (Fig 3).
Lymphocryptovirus BNLF2a Orthologs
The gammaherpesvirus EBV codes for a lytically expressed TAP inhibitor, BNLF2a, that inhibits TAP by interfering with the binding of peptides and ATP to the transporter (Fig 1) [98,99]. Mechanistically, BNLF2a is thought to induce conformational changes in the TAP complex that prevent association of ATP and peptide, but the sequence of events preceding the block in TAP function remains to be elucidated. BNLF2a consists of a hydrophilic N-terminal domain and a hydrophobic C-terminal domain (Fig 2D). BNLF2a lacks an obvious N-terminal signal sequence but is membrane-integrated, nevertheless. The protein was identified as a tail-anchored transmembrane protein that uses Asna1/TRC40, among other proteins, for ER membrane insertion [100]. This mechanism of localizing to membranes is unique among the known viral TAP inhibitors.
Orthologs of BNLF2a have only been identified in lymphocryptoviruses that infect Old World primates [98]. The BNLF2a orthologs encoded by rhesus, chimpanzee, baboon, and gorilla lymphocryptoviruses (RLV, CLV, BLV, and GoLV) share 53%–62% sequence identity with EBV BNLF2a and display a similar disposition of hydrophilic and hydrophobic regions (Fig 2D). When expressed in isolation, these orthologs downregulate cell surface expression of MHC I molecules, indicating conserved TAP-inhibiting properties for BNLF2a proteins expressed by lymphocryptoviruses of Old World primates [98]. No orthologs of BNLF2a have been detected in members of the genus Lymphocryptovirus that infect New World primates, suggesting that the BNLF2a gene was acquired after the divergence of Old World and New World primate lymphocryptoviruses (Fig 3).
Conclusions
Members of all three subfamilies in the family Herpesviridae appear to exploit inhibition of TAP-mediated peptide transport as an immune evasion strategy. The TAP inhibitors that have been identified so far exhibit substantial variation in structural characteristics as well as in mechanisms of action. Yet, despite their large diversity, all inhibitors have evolved to serve a common end: diminish the supply of viral antigenic peptides into the ER lumen in order to avoid elimination of virus-infected cells by MHC I-restricted CTLs, thus ultimately aiding virus replication and spread. The manner in which this has been achieved represents a striking example of functional convergent evolution, and identifies TAP as an Achilles’ heel of the immune system.
All of the TAP-inhibiting proteins described in this review are expressed early during the viral replication cycle [89,91,99,101,102]. For example, EBV BNLF2a is expressed during the (immediate-) early phase of lytic replication, but the protein levels are reduced at later times in infection when other EBV-encoded immune evasive molecules are effective [103,104]. Similarly, BoHV-1 UL49.5 is expressed with early kinetics, but remains present during the late stage of infection [89]. This prolonged expression can be explained by the dual role that UL49.5 plays during viral replication: early in infection, in the absence of gM, it acts as a TAP inhibitor, while at later times of infection it also functions as a chaperone for the late, structural protein gM that facilitates cell-to-cell spread of virus [89]. The early expression of herpesvirus-encoded TAP inhibitors ensures inhibition of the transport of viral peptides into the ER for MHC association shortly after initiation of virus replication, before abundant viral protein synthesis starts. In support of this reasoning, T cell recognition of antigenic peptides expressed early after EBV reactivation is restored in cells infected with a BNLF2a-deleted recombinant EBV [103]. These findings further substantiate the contribution of the virus-encoded TAP inhibitors to immune evasion during infection.
Directly addressing the in vivo relevance of TAP inhibition for human herpesviruses is difficult, because of the restricted host specificity of these viruses. HSV is an exception to the rule, as this virus can productively infect mice. The functional relevance of ICP47 was addressed using a murine ocular infection model. Mice that received uniocular corneal infections with wild-type HSV-1 developed encephalitis and died within 12 days [105]. However, mice infected with an ICP47 deletion mutant did not develop encephalitis, pointing towards a role for ICP47 in preventing the activation of CD8+ T cells that avert the development of lethal encephalitis [105]. An additional study showed that in systemically infected mice, an ICP47 deletion mutant was also attenuated compared to wild type HSV-1. However, in mice lacking TAP, this phenotype was reverted in neuronal tissues, including the brain, suggesting that TAP inhibition is crucial for neuronal infection by HSV-1 [106]. The role of ICP47 in these mouse models seems contradictory to studies showing that ICP47 fails to inhibit TAP efficiently in mouse cell lines [69,70,79,107,108]. The low level of inhibition observed in vitro may be sufficient to generate a phenotype in vivo. Alternatively, ICP47 may block in vivo CD8+ T cell responses by additional mechanisms. In vivo studies on all other human herpesviruses depend on humanized mouse models. The identification of functional orthologs of, for example, US6 and BNLF2a in the genomes of Old World primate-infecting herpesviruses offers new opportunities to evaluate their contribution to replication and spread of the viruses in vivo. RhCMV, which encodes functional homologs of HCMV US2, US3, US6 and US11 [109], has been used to study the role of these immune evasion proteins in vivo [110]. RhCMV is known to be able to reinfect or superinfect its host, despite the presence of high levels of neutralizing antibodies and RhCMV-specific CD4+ and CD8+ T cells. However, rhesus macaques could not be superinfected by a RhCMV strain in which the genetic region encoding the US2, US3, US6, and US11 homologs was deleted. In contrast, the presence of these genes was not required for persistent infection of RhCMV-naïve hosts or for superinfection of macaques transiently depleted of CD8+ T cells. These studies indicate that impairment of MHC I presentation is critical for evading CD8+ T cell responses during superinfection by RhCMV, but not during primary infection [110].
The recent identification of the first viral TAP inhibitor outside the family Herpesviridae highlights the importance of targeting TAP function as a general viral immunoevasive strategy. Poxviruses, like herpesviruses, are known for their elaborate strategies aimed at evading the immune system. Cowpox virus (CPXV) is the first non-herpesvirus that has been found to encode a TAP inhibitor. This protein, CPXV012, is an ER-resident type II transmembrane protein of 69 amino acid residues that inhibits TAP through its ER-luminal domain by interfering with ATP binding to the NBDs of TAP [111–113]. A recent analysis of this gene in a range of CPXV strains has provided interesting clues as to the possible origin of the TAP-inhibiting capacity of CPXV012. It appears to have originated from a frameshifting deletion from a longer protein that possesses an extended ER-luminal region containing a C-type lectin-like domain [113,114]. The longer protein does not block TAP. The majority of the recently isolated clinical strains encode the shorter, TAP-inhibiting protein [113].
The identification of TAP inhibitors in herpesviruses and poxviruses suggests that DNA viruses in particular benefit from interference with TAP function as a means to evade CD8+ T cell responses. The absence of such evasion mechanisms in RNA viruses may in part be explained by the high mutation rate of RNA virus genomes, which allows for CD8+ T cell evasion by antigenic variation [115,116]. The generally lower mutation rate of DNA viruses may require alternative immune evasion mechanisms, including TAP inhibition. The relatively large genomes of herpes - and poxviruses have the capacity to accommodate dedicated immune evasion proteins that counteract the host immune response.
Apart from TAP, MHC I itself is also targeted directly by a wide range of viral proteins encoded by herpesviruses and poxviruses. Among the alphaherpesviruses, ORF66 of VZV binds to and accumulates MHC I in the Golgi compartment [117,118]. Several betaherpesviruses express proteins that induce degradation of MHC I via various pathways. US2, US10, and US11 of HCMV target MHC I for degradation via the ubiquitin-proteasome pathway [27–29]. MCMV-encoded gp48 and U21 of HHV-6 and HHV-7 mediate degradation of MHC I via the endolysosomal route [30,119,120]. The m152 protein of MCMV retains MHC I in intracellular compartments [35]. Rhesus CMV protein rh178 inhibits MHC I heavy chain expression by preventing its co-translational insertion into the ER membrane [121]. Also members of the gammaherpesvirus subfamily induce MHC I degradation using a variety of strategies. EBV-encoded BILF1 reroutes MHC I and targets it for lysosomal degradation [36,122]. MHV-68-encoded mK3 targets MHC I for proteasomal degradation [123], whereas kK3 and kk5 of KSHV degrade MHC I through the endolysosomal pathway [38].
In addition to herperviruses, poxviruses and adenoviruses also encode gene products that target MHC I directly. CPXV203 of cowpox virus and E3-19K of adenovirus both cause retention of MHC I in the ER [124–126].
The reduced overall MHC I surface expression induced by viral inhibitors makes cells more vulnerable for NK cell recognition. NK cells sense the lack of MHC I on target cells; this may result in a response depending on additional activation and inhibitory signals (reviewed by [127]). Herpes - and poxviruses use a variety of gene products to counteract NK cell responses. HCMV has been especially well studied for its ability to evade NK cell-mediated cytotoxicity (reviewed in [128]), but other herpesviruses and poxviruses also encode gene products to counteract NK cell-mediated cytotoxicity [129–131].
In conclusion, four classes of herpesvirus-encoded TAP inhibitors have been identified so far. These appear to be unrelated and to have been acquired independently and relatively recently during evolution, providing a powerful illustration of functional convergent evolution. In addition, poxvirus CPXV012 has been shown to code for yet another type of TAP inhibitor. Where in vitro and in vivo studies have been possible, they have demonstrated the significance of TAP inhibitors in the evasion of CTL recognition and replication of the virus in the face of potent immune responses. In addition to inhibition of TAP, MHC I function is inhibited by a large repertoire of unrelated viral gene products directly targeting MHC I, representing yet another example of functional convergent evolution. The acquisition of a wide range of unrelated proteins that interfere with MHC I-restricted antigen presentation highlights the importance of CTLs in antiviral immunity.
Zdroje
1. McGeoch DJ, Gatherer D (2005) Integrating reptilian herpesviruses into the family herpesviridae. J Virol 79 : 725–731. 15613300
2. Reits EA, Vos JC, Gromme M, Neefjes J (2000) The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature 404 : 774–778. 10783892
3. Princiotta MF, Finzi D, Qian SB, Gibbs J, Schuchmann S, et al. (2003) Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity 18 : 343–354. 12648452
4. Lehner PJ, Surman MJ, Cresswell P (1998) Soluble tapasin restores MHC class I expression and function in the tapasin-negative cell line. 220. Immunity 8 : 221–231. 9492003
5. Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P (1996) Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity 5 : 103–114. 8769474
6. Wearsch PA, Cresswell P (2007) Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nat Immunol 8 : 873–881. 17603487
7. Kienast A, Preuss M, Winkler M, Dick TP (2007) Redox regulation of peptide receptivity of major histocompatibility complex class I molecules by ERp57 and tapasin. Nat Immunol 8 : 864–872. 17603488
8. Koch J, Guntrum R, Tampe R (2006) The first N-terminal transmembrane helix of each subunit of the antigenic peptide transporter TAP is essential for independent tapasin binding. FEBS Lett 580 : 4091–4096. 16828748
9. Leonhardt RM, Keusekotten K, Bekpen C, Knittler MR (2005) Critical role for the tapasin-docking site of TAP2 in the functional integrity of the MHC class I-peptide-loading complex. J Immunol 175 : 5104–5114. 16210614
10. Peaper DR, Wearsch PA, Cresswell P (2005) Tapasin and ERp57 form a stable disulfide-linked dimer within the MHC class I peptide-loading complex. EMBO J 24 : 3613–3623. 16193070
11. Raghavan M, Del Cid N, Rizvi SM, Peters LR (2008) MHC class I assembly: out and about. Trends Immunol 29 : 436–443. doi: 10.1016/j.it.2008.06.004 18675588
12. Wearsch PA, Cresswell P (2008) The quality control of MHC class I peptide loading. Curr Opin Cell Biol 20 : 624–631. doi: 10.1016/j.ceb.2008.09.005 18926908
13. Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, et al. (2009) The order Herpesvirales. Arch Virol 154 : 171–177. doi: 10.1007/s00705-008-0278-4 19066710
14. Staras SA, Dollard SC, Radford KW, Flanders WD, Pass RF, et al. (2006) Seroprevalence of cytomegalovirus infection in the United States, 1988–1994. Clin Infect Dis 43 : 1143–1151. 17029132
15. Kilgore PE, Kruszon-Moran D, Seward JF, Jumaan A, Van Loon FP, et al. (2003) Varicella in Americans from NHANES III: implications for control through routine immunization. J Med Virol 70 Suppl 1: S111–118. 12627498
16. Epstein MA, Achong BG, Barr YM (1964) VIRUS PARTICLES IN CULTURED LYMPHOBLASTS FROM BURKITT'S LYMPHOMA. Lancet 1 : 702–703. 14107961
17. Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, et al. (1994) Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266 : 1865–1869. 7997879
18. Kenneson A, Cannon MJ (2007) Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol 17 : 253–276. 17579921
19. Ressing ME, Horst D, Griffin BD, Tellam J, Zuo J, et al. (2008) Epstein-Barr virus evasion of CD8(+) and CD4(+) T cell immunity via concerted actions of multiple gene products. Semin Cancer Biol 18 : 397–408. doi: 10.1016/j.semcancer.2008.10.008 18977445
20. Liang C, Lee JS, Jung JU (2008) Immune evasion in Kaposi's sarcoma-associated herpes virus associated oncogenesis. Semin Cancer Biol 18 : 423–436. doi: 10.1016/j.semcancer.2008.09.003 18948197
21. Middeldorp JM, Pegtel DM (2008) Multiple roles of LMP1 in Epstein-Barr virus induced immune escape. Semin Cancer Biol 18 : 388–396. doi: 10.1016/j.semcancer.2008.10.004 19013244
22. Fenwick ML, Clark J (1982) Early and delayed shut-off of host protein synthesis in cells infected with herpes simplex virus. J Gen Virol 61 (Pt l): 121–125. 6288847
23. Kwong AD, Frenkel N (1987) Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc Natl Acad Sci U S A 84 : 1926–1930. 3031658
24. Rowe M, Glaunsinger B, van Leeuwen D, Zuo J, Sweetman D, et al. (2007) Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc Natl Acad Sci U S A 104 : 3366–3371. 17360652
25. Zuo J, Thomas W, van Leeuwen D, Middeldorp JM, Wiertz EJ, et al. (2008) The DNase of gammaherpesviruses impairs recognition by virus-specific CD8+ T cells through an additional host shutoff function. J Virol 82 : 2385–2393. 18094150
26. Glaunsinger B, Chavez L, Ganem D (2005) The exonuclease and host shutoff functions of the SOX protein of Kaposi's sarcoma-associated herpesvirus are genetically separable. J Virol 79 : 7396–7401. 15919895
27. Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, et al. (1996) The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84 : 769–779. 8625414
28. Park B, Spooner E, Houser BL, Strominger JL, Ploegh HL (2010) The HCMV membrane glycoprotein US10 selectively targets HLA-G for degradation. J Exp Med 207 : 2033–2041. doi: 10.1084/jem.20091793 20713594
29. Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, et al. (1996) Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384 : 432–438. 8945469
30. Reusch U, Muranyi W, Lucin P, Burgert HG, Hengel H, et al. (1999) A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. EMBO J 18 : 1081–1091. 10022849
31. Lybarger L, Wang X, Harris MR, Virgin HWt, Hansen TH (2003) Virus subversion of the MHC class I peptide-loading complex. Immunity 18 : 121–130. 12530981
32. Stevenson PG, May JS, Smith XG, Marques S, Adler H, et al. (2002) K3-mediated evasion of CD8(+) T cells aids amplification of a latent gamma-herpesvirus. Nat Immunol 3 : 733–740. 12101398
33. Wang X, Herr RA, Chua WJ, Lybarger L, Wiertz EJ, et al. (2007) Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J Cell Biol 177 : 613–624. 17502423
34. Jones TR, Wiertz EJ, Sun L, Fish KN, Nelson JA, et al. (1996) Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc Natl Acad Sci U S A 93 : 11327–11333. 8876135
35. Ziegler H, Thale R, Lucin P, Muranyi W, Flohr T, et al. (1997) A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity 6 : 57–66. 9052837
36. Zuo J, Currin A, Griffin BD, Shannon-Lowe C, Thomas WA, et al. (2009) The Epstein-Barr virus G-protein-coupled receptor contributes to immune evasion by targeting MHC class I molecules for degradation. PLoS Pathog 5: e1000255. doi: 10.1371/journal.ppat.1000255 19119421
37. Coscoy L, Ganem D (2000) Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc Natl Acad Sci U S A 97 : 8051–8056. 10859362
38. Ishido S, Wang C, Lee BS, Cohen GB, Jung JU (2000) Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J Virol 74 : 5300–5309. 10799607
39. Lehner PJ, Hoer S, Dodd R, Duncan LM (2005) Downregulation of cell surface receptors by the K3 family of viral and cellular ubiquitin E3 ligases. Immunol Rev 207 : 112–125. 16181331
40. Hulpke S, Baldauf C, Tampe R (2012) Molecular architecture of the MHC I peptide-loading complex: one tapasin molecule is essential and sufficient for antigen processing. FASEB J 26 : 5071–5080. doi: 10.1096/fj.12-217489 22923333
41. Koch J, Guntrum R, Heintke S, Kyritsis C, Tampe R (2004) Functional dissection of the transmembrane domains of the transporter associated with antigen processing (TAP). J Biol Chem 279 : 10142–10147. 14679198
42. Androlewicz MJ, Cresswell P (1994) Human transporters associated with antigen processing possess a promiscuous peptide-binding site. Immunity 1 : 7–14. 7889401
43. Koopmann JO, Post M, Neefjes JJ, Hammerling GJ, Momburg F (1996) Translocation of long peptides by transporters associated with antigen processing (TAP). Eur J Immunol 26 : 1720–1728. 8765012
44. Momburg F, Roelse J, Hammerling GJ, Neefjes JJ (1994) Peptide size selection by the major histocompatibility complex-encoded peptide transporter. J Exp Med 179 : 1613–1623. 8163941
45. Schumacher TN, Kantesaria DV, Heemels MT, Ashton-Rickardt PG, Shepherd JC, et al. (1994) Peptide length and sequence specificity of the mouse TAP1/TAP2 translocator. J Exp Med 179 : 533–540. 8294864
46. van Endert PM, Tampe R, Meyer TH, Tisch R, Bach JF, et al. (1994) A sequential model for peptide binding and transport by the transporters associated with antigen processing. Immunity 1 : 491–500. 7895159
47. Nijenhuis M, Hammerling GJ (1996) Multiple regions of the transporter associated with antigen processing (TAP) contribute to its peptide binding site. J Immunol 157 : 5467–5477. 8955196
48. Armandola EA, Momburg F, Nijenhuis M, Bulbuc N, Fruh K, et al. (1996) A point mutation in the human transporter associated with antigen processing (TAP2) alters the peptide transport specificity. Eur J Immunol 26 : 1748–1755. 8765016
49. Corradi V, Singh G, Tieleman DP (2012) The human transporter associated with antigen processing: molecular models to describe peptide binding competent states. J Biol Chem 287 : 28099–28111. doi: 10.1074/jbc.M112.381251 22700967
50. Procko E, Ferrin-O'Connell I, Ng SL, Gaudet R (2006) Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Mol Cell 24 : 51–62. 17018292
51. Chen M, Abele R, Tampe R (2003) Peptides induce ATP hydrolysis at both subunits of the transporter associated with antigen processing. J Biol Chem 278 : 29686–29692. 12777379
52. Lapinski PE, Raghuraman G, Raghavan M (2003) Nucleotide interactions with membrane-bound transporter associated with antigen processing proteins. J Biol Chem 278 : 8229–8237. 12501238
53. Perria CL, Rajamanickam V, Lapinski PE, Raghavan M (2006) Catalytic site modifications of TAP1 and TAP2 and their functional consequences. J Biol Chem 281 : 39839–39851. 17068338
54. Saveanu L, Daniel S, van Endert PM (2001) Distinct functions of the ATP binding cassettes of transporters associated with antigen processing: a mutational analysis of Walker A and B sequences. J Biol Chem 276 : 22107–22113. 11290739
55. Dawson RJ, Locher KP (2006) Structure of a bacterial multidrug ABC transporter. Nature 443 : 180–185. 16943773
56. Hohl M, Briand C, Grutter MG, Seeger MA (2012) Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat Struct Mol Biol 19 : 395–402. doi: 10.1038/nsmb.2267 22447242
57. Kim J, Wu S, Tomasiak TM, Mergel C, Winter MB, et al. (2015) Subnanometre-resolution electron cryomicroscopy structure of a heterodimeric ABC exporter. Nature 517 : 396–400. doi: 10.1038/nature13872 25363761
58. Kodan A, Yamaguchi T, Nakatsu T, Sakiyama K, Hipolito CJ, et al. (2014) Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog. Proc Natl Acad Sci U S A 111 : 4049–4054. doi: 10.1073/pnas.1321562111 24591620
59. Srinivasan V, Pierik AJ, Lill R (2014) Crystal structures of nucleotide-free and glutathione-bound mitochondrial ABC transporter Atm1. Science 343 : 1137–1140. doi: 10.1126/science.1246729 24604199
60. Geng J, Sivaramakrishnan S, Raghavan M (2013) Analyses of conformational states of the transporter associated with antigen processing (TAP) protein in a native cellular membrane environment. J Biol Chem 288 : 37039–37047. doi: 10.1074/jbc.M113.504696 24196954
61. Grossmann N, Vakkasoglu AS, Hulpke S, Abele R, Gaudet R, et al. (2014) Mechanistic determinants of the directionality and energetics of active export by a heterodimeric ABC transporter. Nat Commun 5 : 5419. doi: 10.1038/ncomms6419 25377891
62. Mayerhofer PU, Tampe R (2014) Antigen Translocation Machineries in Adaptive Immunity and Viral Immune Evasion. J Mol Biol. 427 : 1102–1118 doi: 10.1016/j.jmb.2014.09.006 25224907
63. Spies T, DeMars R (1991) Restored expression of major histocompatibility class I molecules by gene transfer of a putative peptide transporter. Nature 351 : 323–324. 2034277
64. Spies T, Cerundolo V, Colonna M, Cresswell P, Townsend A, et al. (1992) Presentation of viral antigen by MHC class I molecules is dependent on a putative peptide transporter heterodimer. Nature 355 : 644–646. 1538752
65. de la Salle H, Hanau D, Fricker D, Urlacher A, Kelly A, et al. (1994) Homozygous human TAP peptide transporter mutation in HLA class I deficiency. Science 265 : 237–241. 7517574
66. de la Salle H, Zimmer J, Fricker D, Angenieux C, Cazenave JP, et al. (1999) HLA class I deficiencies due to mutations in subunit 1 of the peptide transporter TAP1. J Clin Invest 103: R9–R13. 10074495
67. Moins-Teisserenc HT, Gadola SD, Cella M, Dunbar PR, Exley A, et al. (1999) Association of a syndrome resembling Wegener's granulomatosis with low surface expression of HLA class-I molecules. Lancet 354 : 1598–1603. 10560675
68. de la Salle H, Saulquin X, Mansour I, Klayme S, Fricker D, et al. (2002) Asymptomatic deficiency in the peptide transporter associated to antigen processing (TAP). Clin Exp Immunol 128 : 525–531. 12067308
69. Ahn K, Meyer TH, Uebel S, Sempe P, Djaballah H, et al. (1996) Molecular mechanism and species specificity of TAP inhibition by herpes simplex virus ICP47. EMBO J 15 : 3247–3255. 8670825
70. Fruh K, Ahn K, Djaballah H, Sempe P, van Endert PM, et al. (1995) A viral inhibitor of peptide transporters for antigen presentation. Nature 375 : 415–418. 7760936
71. Hill A, Jugovic P, York I, Russ G, Bennink J, et al. (1995) Herpes simplex virus turns off the TAP to evade host immunity. Nature 375 : 411–415. 7760935
72. Tomazin R, Hill AB, Jugovic P, York I, van Endert P, et al. (1996) Stable binding of the herpes simplex virus ICP47 protein to the peptide binding site of TAP. EMBO J 15 : 3256–3266. 8670826
73. Beinert D, Neumann L, Uebel S, Tampe R (1997) Structure of the viral TAP-inhibitor ICP47 induced by membrane association. Biochemistry 36 : 4694–4700. 9109681
74. Galocha B, Hill A, Barnett BC, Dolan A, Raimondi A, et al. (1997) The active site of ICP47, a herpes simplex virus-encoded inhibitor of the major histocompatibility complex (MHC)-encoded peptide transporter associated with antigen processing (TAP), maps to the NH2-terminal 35 residues. J Exp Med 185 : 1565–1572. 9151894
75. Neumann L, Kraas W, Uebel S, Jung G, Tampe R (1997) The active domain of the herpes simplex virus protein ICP47: a potent inhibitor of the transporter associated with antigen processing. J Mol Biol 272 : 484–492. 9325106
76. Lacaille VG, Androlewicz MJ (1998) Herpes simplex virus inhibitor ICP47 destabilizes the transporter associated with antigen processing (TAP) heterodimer. J Biol Chem 273 : 17386–17390. 9651323
77. Seyffer F, Tampe R (2015) ABC transporters in adaptive immunity. Biochim Biophys Acta 1850 : 449–460. doi: 10.1016/j.bbagen.2014.05.022 24923865
78. Bigger JE, Martin DW (2004) Identification of an ICP47 homologue in simian agent 8 (SA8). Virus Genes 28 : 223–225. 15077611
79. Tomazin R, van Schoot NE, Goldsmith K, Jugovic P, Sempe P, et al. (1998) Herpes simplex virus type 2 ICP47 inhibits human TAP but not mouse TAP. J Virol 72 : 2560–2563. 9499125
80. Vasireddi M, Hilliard J (2012) Herpes B virus, macacine herpesvirus 1, breaks simplex virus tradition via major histocompatibility complex class I expression in cells from human and macaque hosts. J Virol 86 : 12503–12511. doi: 10.1128/JVI.01350-12 22973043
81. McGeoch DJ, Davison AJ (1999) The molecular evolutionary history of the herpesviruses.; Domingo E, Webster R, Holland J, editors. London: Academic Press. 441–465 p.
82. Koppers-Lalic D, Reits EA, Ressing ME, Lipinska AD, Abele R, et al. (2005) Varicelloviruses avoid T cell recognition by UL49.5-mediated inactivation of the transporter associated with antigen processing. Proc Natl Acad Sci U S A 102 : 5144–5149. 15793001
83. Verweij MC, Lipinska AD, Koppers-Lalic D, Quinten E, Funke J, et al. (2011) Structural and functional analysis of the TAP-inhibiting UL49.5 proteins of varicelloviruses. Mol Immunol 48 : 2038–2051. doi: 10.1016/j.molimm.2011.06.438 21764135
84. Koppers-Lalic D, Verweij MC, Lipinska AD, Wang Y, Quinten E, et al. (2008) Varicellovirus UL 49.5 proteins differentially affect the function of the transporter associated with antigen processing, TAP. PLoS Pathog 4: e1000080. doi: 10.1371/journal.ppat.1000080 18516302
85. Verweij MC, Lipinska AD, Koppers-Lalic D, van Leeuwen WF, Cohen JI, et al. (2011) The capacity of UL49.5 proteins to inhibit TAP is widely distributed among members of the genus Varicellovirus. J Virol 85 : 2351–2363. doi: 10.1128/JVI.01621-10 21159875
86. Jons A, Dijkstra JM, Mettenleiter TC (1998) Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J Virol 72 : 550–557. 9420258
87. Rudolph J, Seyboldt C, Granzow H, Osterrieder N (2002) The gene 10 (UL49.5) product of equine herpesvirus 1 is necessary and sufficient for functional processing of glycoprotein M. J Virol 76 : 2952–2963. 11861861
88. Wu SX, Zhu XP, Letchworth GJ (1998) Bovine herpesvirus 1 glycoprotein M forms a disulfide-linked heterodimer with the U(L)49.5 protein. J Virol 72 : 3029–3036. 9525625
89. Lipinska AD, Koppers-Lalic D, Rychlowski M, Admiraal P, Rijsewijk FA, et al. (2006) Bovine herpesvirus 1 UL49.5 protein inhibits the transporter associated with antigen processing despite complex formation with glycoprotein M. J Virol 80 : 5822–5832. 16731921
90. Ahn K, Gruhler A, Galocha B, Jones TR, Wiertz EJ, et al. (1997) The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6 : 613–621. 9175839
91. Hengel H, Koopmann JO, Flohr T, Muranyi W, Goulmy E, et al. (1997) A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity 6 : 623–632. 9175840
92. Lehner PJ, Karttunen JT, Wilkinson GW, Cresswell P (1997) The human cytomegalovirus US6 glycoprotein inhibits transporter associated with antigen processing-dependent peptide translocation. Proc Natl Acad Sci U S A 94 : 6904–6909. 9192664
93. Hewitt EW, Gupta SS, Lehner PJ (2001) The human cytomegalovirus gene product US6 inhibits ATP binding by TAP. EMBO J 20 : 387–396. 11157746
94. van Endert PM, Saveanu L, Hewitt EW, Lehner P (2002) Powering the peptide pump: TAP crosstalk with energetic nucleotides. Trends Biochem Sci 27 : 454–461. 12217520
95. Halenius A, Momburg F, Reinhard H, Bauer D, Lobigs M, et al. (2006) Physical and functional interactions of the cytomegalovirus US6 glycoprotein with the transporter associated with antigen processing. J Biol Chem 281 : 5383–5390. 16356928
96. Rawlinson WD, Farrell HE, Barrell BG (1996) Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol 70 : 8833–8849. 8971012
97. Davison AJ, Holton M, Dolan A, Dargan DJ, Gatherer D, et al. (2013) Comparative Genomics of Primate Cytomegaloviruses; Reddehase MJ, editor. Norwich, UK: Caister Academic Press.
98. Hislop AD, Ressing ME, van Leeuwen D, Pudney VA, Horst D, et al. (2007) A CD8+ T cell immune evasion protein specific to Epstein-Barr virus and its close relatives in Old World primates. J Exp Med 204 : 1863–1873. 17620360
99. Horst D, van Leeuwen D, Croft NP, Garstka MA, Hislop AD, et al. (2009) Specific targeting of the EBV lytic phase protein BNLF2a to the transporter associated with antigen processing results in impairment of HLA class I-restricted antigen presentation. J Immunol 182 : 2313–2324. doi: 10.4049/jimmunol.0803218 19201886
100. Horst D, Favaloro V, Vilardi F, van Leeuwen HC, Garstka MA, et al. (2011) EBV protein BNLF2a exploits host tail-anchored protein integration machinery to inhibit TAP. J Immunol 186 : 3594–3605. doi: 10.4049/jimmunol.1002656 21296983
101. York IA, Roop C, Andrews DW, Riddell SR, Graham FL, et al. (1994) A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell 77 : 525–535. 8187174
102. Yuan J, Cahir-McFarland E, Zhao B, Kieff E (2006) Virus and cell RNAs expressed during Epstein-Barr virus replication. J Virol 80 : 2548–2565. 16474161
103. Croft NP, Shannon-Lowe C, Bell AI, Horst D, Kremmer E, et al. (2009) Stage-specific inhibition of MHC class I presentation by the Epstein-Barr virus BNLF2a protein during virus lytic cycle. PLoS Pathog 5: e1000490. doi: 10.1371/journal.ppat.1000490 19557156
104. Quinn LL, Zuo J, Abbott RJ, Shannon-Lowe C, Tierney RJ, et al. (2014) Cooperation between Epstein-Barr virus immune evasion proteins spreads protection from CD8+ T cell recognition across all three phases of the lytic cycle. PLoS Pathog 10: e1004322. doi: 10.1371/journal.ppat.1004322 25144360
105. Goldsmith K, Chen W, Johnson DC, Hendricks RL (1998) Infected cell protein (ICP)47 enhances herpes simplex virus neurovirulence by blocking the CD8+ T cell response. J Exp Med 187 : 341–348. 9449714
106. Burgos JS, Serrano-Saiz E, Sastre I, Valdivieso F (2006) ICP47 mediates viral neuroinvasiveness by induction of TAP protein following intravenous inoculation of herpes simplex virus type 1 in mice. J Neurovirol 12 : 420–427. 17162658
107. Jugovic P, Hill AM, Tomazin R, Ploegh H, Johnson DC (1998) Inhibition of major histocompatibility complex class I antigen presentation in pig and primate cells by herpes simplex virus type 1 and 2 ICP47. J Virol 72 : 5076–5084. 9573278
108. Verweij MC, Ressing ME, Knetsch W, Quinten E, Halenius A, et al. (2011) Inhibition of mouse TAP by immune evasion molecules encoded by non-murine herpesviruses. Mol Immunol 48 : 835–845. doi: 10.1016/j.molimm.2010.12.008 21292324
109. Pande NT, Powers C, Ahn K, Fruh K (2005) Rhesus cytomegalovirus contains functional homologues of US2, US3, US6, and US11. J Virol 79 : 5786–5798. 15827193
110. Hansen SG, Powers CJ, Richards R, Ventura AB, Ford JC, et al. (2010) Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science 328 : 102–106. doi: 10.1126/science.1185350 20360110
111. Alzhanova D, Edwards DM, Hammarlund E, Scholz IG, Horst D, et al. (2009) Cowpox virus inhibits the transporter associated with antigen processing to evade T cell recognition. Cell Host Microbe 6 : 433–445. doi: 10.1016/j.chom.2009.09.013 19917498
112. Byun M, Verweij MC, Pickup DJ, Wiertz EJ, Hansen TH, et al. (2009) Two mechanistically distinct immune evasion proteins of cowpox virus combine to avoid antiviral CD8 T cells. Cell Host Microbe 6 : 422–432. doi: 10.1016/j.chom.2009.09.012 19917497
113. Luteijn RD, Hoelen H, Kruse E, van Leeuwen WF, Grootens J, et al. (2014) Cowpox Virus Protein CPXV012 Eludes CTLs by Blocking ATP Binding to TAP. J Immunol. 193 : 1578–89. doi: 10.4049/jimmunol.1400964 25024387
114. Lin J, Eggensperger S, Hank S, Wycisk AI, Wieneke R, et al. (2014) A negative feedback modulator of antigen processing evolved from a frameshift in the cowpox virus genome. PLoS Pathog 10: e1004554. doi: 10.1371/journal.ppat.1004554 25503639
115. Elena SF, Sanjuan R (2005) Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J Virol 79 : 11555–11558. 16140732
116. Finlay BB, McFadden G (2006) Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124 : 767–782. 16497587
117. Abendroth A, Lin I, Slobedman B, Ploegh H, Arvin AM (2001) Varicella-zoster virus retains major histocompatibility complex class I proteins in the Golgi compartment of infected cells. J Virol 75 : 4878–4888. 11312359
118. Eisfeld AJ, Yee MB, Erazo A, Abendroth A, Kinchington PR (2007) Downregulation of class I major histocompatibility complex surface expression by varicella-zoster virus involves open reading frame 66 protein kinase-dependent and-independent mechanisms. J Virol 81 : 9034–9049. 17567702
119. Glosson NL, Gonyo P, May NA, Schneider CL, Ristow LC, et al. (2010) Insight into the mechanism of human herpesvirus 7 U21-mediated diversion of class I MHC molecules to lysosomes. J Biol Chem 285 : 37016–37029. doi: 10.1074/jbc.M110.125849 20833720
120. Glosson NL, Hudson AW (2007) Human herpesvirus-6A and -6B encode viral immunoevasins that downregulate class I MHC molecules. Virology 365 : 125–135. 17467766
121. Powers CJ, Fruh K (2008) Signal peptide-dependent inhibition of MHC class I heavy chain translation by rhesus cytomegalovirus. PLoS Pathog 4: e1000150. doi: 10.1371/journal.ppat.1000150 18833297
122. Zuo J, Quinn LL, Tamblyn J, Thomas WA, Feederle R, et al. (2011) The Epstein-Barr virus-encoded BILF1 protein modulates immune recognition of endogenously processed antigen by targeting major histocompatibility complex class I molecules trafficking on both the exocytic and endocytic pathways. J Virol 85 : 1604–1614. doi: 10.1128/JVI.01608-10 21123379
123. Boname JM, Stevenson PG (2001) MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15 : 627–636. 11672544
124. Byun M, Wang X, Pak M, Hansen TH, Yokoyama WM (2007) Cowpox virus exploits the endoplasmic reticulum retention pathway to inhibit MHC class I transport to the cell surface. Cell Host Microbe 2 : 306–315. 18005752
125. Cox JH, Yewdell JW, Eisenlohr LC, Johnson PR, Bennink JR (1990) Antigen presentation requires transport of MHC class I molecules from the endoplasmic reticulum. Science 247 : 715–718. 2137259
126. McCoy WHt, Wang X, Yokoyama WM, Hansen TH, Fremont DH (2012) Structural mechanism of ER retrieval of MHC class I by cowpox. PLoS Biol 10: e1001432. doi: 10.1371/journal.pbio.1001432 23209377
127. Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S (2013) Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol 31 : 227–258. doi: 10.1146/annurev-immunol-020711-075005 23516982
128. Wilkinson GW, Tomasec P, Stanton RJ, Armstrong M, Prod'homme V, et al. (2008) Modulation of natural killer cells by human cytomegalovirus. J Clin Virol 41 : 206–212. 18069056
129. Campbell JA, Trossman DS, Yokoyama WM, Carayannopoulos LN (2007) Zoonotic orthopoxviruses encode a high-affinity antagonist of NKG2D. J Exp Med 204 : 1311–1317. 17548517
130. Grauwet K, Cantoni C, Parodi M, De Maria A, Devriendt B, et al. (2014) Modulation of CD112 by the alphaherpesvirus gD protein suppresses DNAM-1-dependent NK cell-mediated lysis of infected cells. Proc Natl Acad Sci U S A 111 : 16118–16123. doi: 10.1073/pnas.1409485111 25352670
131. Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O (2009) Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5 : 376–385. doi: 10.1016/j.chom.2009.03.003 19380116
132. Verweij MC, Koppers-Lalic D, Loch S, Klauschies F, de la Salle H, et al. (2008) The varicellovirus UL49.5 protein blocks the transporter associated with antigen processing (TAP) by inhibiting essential conformational transitions in the 6+6 transmembrane TAP core complex. J Immunol 181 : 4894–4907. 18802093
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Relay of Herpes Simplex Virus between Langerhans Cells and Dermal Dendritic Cells in Human SkinČlánek Does the Arthropod Microbiota Impact the Establishment of Vector-Borne Diseases in Mammalian Hosts?Článek The Ebola Epidemic Crystallizes the Potential of Passive Antibody Therapy for Infectious DiseasesČlánek Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting PolypeptideČlánek A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 4- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Pathogens as Biological Weapons of Invasive Species
- Selection and Spread of Artemisinin-Resistant Alleles in Thailand Prior to the Global Artemisinin Resistance Containment Campaign
- Endopeptidase-Mediated Beta Lactam Tolerance
- Prospective Large-Scale Field Study Generates Predictive Model Identifying Major Contributors to Colony Losses
- Relay of Herpes Simplex Virus between Langerhans Cells and Dermal Dendritic Cells in Human Skin
- Structural Determinants of Phenotypic Diversity and Replication Rate of Human Prions
- Sigma Factor SigB Is Crucial to Mediate Adaptation during Chronic Infections
- EphrinA2 Receptor (EphA2) Is an Invasion and Intracellular Signaling Receptor for
- Toxin-Induced Necroptosis Is a Major Mechanism of Lung Damage
- Heterologous Expression in Remodeled . : A Platform for Monoaminergic Agonist Identification and Anthelmintic Screening
- Novel Disease Susceptibility Factors for Fungal Necrotrophic Pathogens in Arabidopsis
- Interleukin 21 Signaling in B Cells Is Required for Efficient Establishment of Murine Gammaherpesvirus Latency
- Phosphorylation at the Homotypic Interface Regulates Nucleoprotein Oligomerization and Assembly of the Influenza Virus Replication Machinery
- Human Papillomaviruses Activate and Recruit SMC1 Cohesin Proteins for the Differentiation-Dependent Life Cycle through Association with CTCF Insulators
- Ubiquitous Promoter-Localization of Essential Virulence Regulators in
- TGF-β Suppression of HBV RNA through AID-Dependent Recruitment of an RNA Exosome Complex
- The Immune Adaptor ADAP Regulates Reciprocal TGF-β1-Integrin Crosstalk to Protect from Influenza Virus Infection
- Antagonism of miR-328 Increases the Antimicrobial Function of Macrophages and Neutrophils and Rapid Clearance of Non-typeable (NTHi) from Infected Lung
- The Epigenetic Regulator G9a Mediates Tolerance to RNA Virus Infection in
- Does the Arthropod Microbiota Impact the Establishment of Vector-Borne Diseases in Mammalian Hosts?
- Hantaan Virus Infection Induces Both Th1 and ThGranzyme B+ Cell Immune Responses That Associated with Viral Control and Clinical Outcome in Humans
- Viral Inhibition of the Transporter Associated with Antigen Processing (TAP): A Striking Example of Functional Convergent Evolution
- Plasma Membrane Profiling Defines an Expanded Class of Cell Surface Proteins Selectively Targeted for Degradation by HCMV US2 in Cooperation with UL141
- Optineurin Regulates the Interferon Response in a Cell Cycle-Dependent Manner
- IFIT1 Differentially Interferes with Translation and Replication of Alphavirus Genomes and Promotes Induction of Type I Interferon
- The EBNA3 Family of Epstein-Barr Virus Nuclear Proteins Associates with the USP46/USP12 Deubiquitination Complexes to Regulate Lymphoblastoid Cell Line Growth
- Hepatitis C Virus RNA Replication Depends on Specific and -Acting Activities of Viral Nonstructural Proteins
- A Neuron-Specific Antiviral Mechanism Prevents Lethal Flaviviral Infection of Mosquitoes
- The Aspartate-Less Receiver (ALR) Domains: Distribution, Structure and Function
- Global Genome and Transcriptome Analyses of Epidemic Isolate 98-06 Uncover Novel Effectors and Pathogenicity-Related Genes, Revealing Gene Gain and Lose Dynamics in Genome Evolution
- The Ebola Epidemic Crystallizes the Potential of Passive Antibody Therapy for Infectious Diseases
- Ebola Virus Entry: A Curious and Complex Series of Events
- Conserved Spirosomes Suggest a Single Type of Transformation Pilus in Competence
- Spatial Structure, Transmission Modes and the Evolution of Viral Exploitation Strategies
- Bacterial Cooperation Causes Systematic Errors in Pathogen Risk Assessment due to the Failure of the Independent Action Hypothesis
- Transgenic Fatal Familial Insomnia Mice Indicate Prion Infectivity-Independent Mechanisms of Pathogenesis and Phenotypic Expression of Disease
- Cerebrospinal Fluid Cytokine Profiles Predict Risk of Early Mortality and Immune Reconstitution Inflammatory Syndrome in HIV-Associated Cryptococcal Meningitis
- Utilize Host Actin for Efficient Maternal Transmission in
- Borna Disease Virus Phosphoprotein Impairs the Developmental Program Controlling Neurogenesis and Reduces Human GABAergic Neurogenesis
- An Effector Peptide Family Required for Toll-Mediated Immunity
- Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting Polypeptide
- A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development
- Quadruple Quorum-Sensing Inputs Control Virulence and Maintain System Robustness
- Leukocyte-Derived IFN-α/β and Epithelial IFN-λ Constitute a Compartmentalized Mucosal Defense System that Restricts Enteric Virus Infections
- A Strategy for O-Glycoproteomics of Enveloped Viruses—the O-Glycoproteome of Herpes Simplex Virus Type 1
- Macrocyclic Lactones Differ in Interaction with Recombinant P-Glycoprotein 9 of the Parasitic Nematode and Ketoconazole in a Yeast Growth Assay
- Neofunctionalization of the α1,2fucosyltransferase Paralogue in Leporids Contributes to Glycan Polymorphism and Resistance to Rabbit Hemorrhagic Disease Virus
- The Extracytoplasmic Linker Peptide of the Sensor Protein SaeS Tunes the Kinase Activity Required for Staphylococcal Virulence in Response to Host Signals
- Murine CMV-Induced Hearing Loss Is Associated with Inner Ear Inflammation and Loss of Spiral Ganglia Neurons
- Dual miRNA Targeting Restricts Host Range and Attenuates Neurovirulence of Flaviviruses
- GATA-Dependent Glutaminolysis Drives Appressorium Formation in by Suppressing TOR Inhibition of cAMP/PKA Signaling
- Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Infection
- Genetic Analysis Using an Isogenic Mating Pair of Identifies Azole Resistance Genes and Lack of Locus’s Role in Virulence
- A Temporal Gate for Viral Enhancers to Co-opt Toll-Like-Receptor Transcriptional Activation Pathways upon Acute Infection
- Neutrophil Recruitment to Lymph Nodes Limits Local Humoral Response to
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Toxin-Induced Necroptosis Is a Major Mechanism of Lung Damage
- Transgenic Fatal Familial Insomnia Mice Indicate Prion Infectivity-Independent Mechanisms of Pathogenesis and Phenotypic Expression of Disease
- Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Infection
- EphrinA2 Receptor (EphA2) Is an Invasion and Intracellular Signaling Receptor for
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



