-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaFunctional Variants in and Involved in Activation of the NF-κB Pathway Are Associated with Rheumatoid Arthritis in Japanese
Rheumatoid arthritis is an autoimmune disease with a complex etiology, leading to inflammation of synovial tissue and joint destruction. Through a genome-wide association study (GWAS) and two replication studies in the Japanese population (7,907 cases and 35,362 controls), we identified two gene loci associated with rheumatoid arthritis susceptibility (NFKBIE at 6p21.1, rs2233434, odds ratio (OR) = 1.20, P = 1.3×10−15; RTKN2 at 10q21.2, rs3125734, OR = 1.20, P = 4.6×10−9). In addition to two functional non-synonymous SNPs in NFKBIE, we identified candidate causal SNPs with regulatory potential in NFKBIE and RTKN2 gene regions by integrating in silico analysis using public genome databases and subsequent in vitro analysis. Both of these genes are known to regulate the NF-κB pathway, and the risk alleles of the genes were implicated in the enhancement of NF-κB activity in our analyses. These results suggest that the NF-κB pathway plays a role in pathogenesis and would be a rational target for treatment of rheumatoid arthritis.
Published in the journal: . PLoS Genet 8(9): e32767. doi:10.1371/journal.pgen.1002949
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002949Summary
Rheumatoid arthritis is an autoimmune disease with a complex etiology, leading to inflammation of synovial tissue and joint destruction. Through a genome-wide association study (GWAS) and two replication studies in the Japanese population (7,907 cases and 35,362 controls), we identified two gene loci associated with rheumatoid arthritis susceptibility (NFKBIE at 6p21.1, rs2233434, odds ratio (OR) = 1.20, P = 1.3×10−15; RTKN2 at 10q21.2, rs3125734, OR = 1.20, P = 4.6×10−9). In addition to two functional non-synonymous SNPs in NFKBIE, we identified candidate causal SNPs with regulatory potential in NFKBIE and RTKN2 gene regions by integrating in silico analysis using public genome databases and subsequent in vitro analysis. Both of these genes are known to regulate the NF-κB pathway, and the risk alleles of the genes were implicated in the enhancement of NF-κB activity in our analyses. These results suggest that the NF-κB pathway plays a role in pathogenesis and would be a rational target for treatment of rheumatoid arthritis.
Introduction
Rheumatoid arthritis (RA [MIM 180300]) is an autoimmune disease [1] with a complex etiology involving several genetic factors as well as environmental factors. Previous genome-wide association studies (GWAS) for RA have discovered many genetic loci [2]–[6], although the causal mechanisms linking the variants in these loci and disease etiology are largely unknown, except for in a few cases [6]–[8]. In contrast to mutations in Mendelian, monogenic diseases, most disease-associated variants in complex diseases, including autoimmune diseases, have moderate effects on disease susceptibility. This is because the disease causal variants in complex diseases are thought to have moderate effects on gene function, while amino acid changes introduced by the mutations of monogenic diseases have critical impacts on protein function [9]. Moreover, it has been demonstrated that the majority of autoimmune disease loci are expression quantitative trait loci (eQTLs) [10], [11], indicating that accumulation of quantitative, but not qualitative, changes in gene function likely predisposes individuals to the disease. This renders it difficult to pinpoint the causal variants in the GWAS loci, especially in eQTLs, because all the variations in strong linkage disequilibrium (LD) with the marker SNP in a GWAS, the majority of which are not covered by the SNP array, are possible candidates for causal variants.
In recent years, with the emergence of next-generation sequencing technologies, the way we approach disease-causing variants has dramatically changed. First, a comprehensive map of human genetic variations is now available owing to the 1000 Genome Project [12], which allows us to grasp most of the potential common variants. This also enables us to perform genotype imputation of SNPs that are not directly genotyped in the GWAS, and consequently, to test them for association. Second, genomic studies using new technologies, such as chromatin immunoprecipitation-sequencing (ChIP-seq) and DNase I hypersensitive sites sequencing (DNase-seq), have advanced our understanding of how each genomic cluster regulates gene transcription. If disease-associated variants are present in a critical site for gene regulation suggested by the ChIP-seq and DNase-seq studies, the disease-associated variants might possibly influence gene transcription levels such as through altered transcription factor-DNA binding avidity.
In the present study, we first performed replication studies of candidate loci in our previous GWAS and identified two association signals with genome-wide significance (P<5×10−8) in nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon (NFKBIE [MIM 604548]) and rhotekin 2 (RTKN2) loci. By utilizing publicly available datasets yielded by the above-mentioned genomic studies, we then performed integrated in silico and in vitro analysis to identify plausible causal variants in NFKBIE and RTKN2 loci.
Results
Identification of rheumatoid arthritis susceptibility genes
We previously performed a GWAS of RA using a Japanese case-control cohort (2,303 cases and 3,380 controls) and identified significant associations in major histocompatibility complex, class II, DR beta 1 (HLA-DRB1 [MIM 142857]), and chemokine (C-C motif) receptor 6 (CCR6 [MIM 601835]) loci (PGWAS<5×10−8) [6]. To reveal additional risk loci from those showing moderate associations in the GWAS (31 loci, 5×10−8<PGWAS<5×10−5), we selected a landmark SNP from each locus and genotyped it for an additional cohort (replication-1 : 2,187 cases and 28,219 controls) (Table S1, S2). Among the 31 SNPs genotyped, seven SNPs were nominally associated with RA (P<0.05), which included SNPs in the tumor necrosis factor, alpha-induced protein 3 (TNFAIP3 [MIM 191163]), and signal transducer and the activator of transcription 4 (STAT4 [MIM 600558]) gene loci that were previously reported to be associated with RA [13], [14] (Table S2). In a combined analysis of the GWAS and the 1st replication study, we identified two associations with genome-wide significance (P<5×10−8) in NFKBIE (6p21.1, rs2233434, P = 4.1×10−11, odds ratio (OR) = 1.21, 95% confidence interval (CI) = 1.14–1.28) and in RTKN2 (10q21.2, rs3125734, P = 3.7×10−8, OR = 1.23, 95% CI = 1.14–1.32) (Table 1 and Figure 1). NFKBIE was previously reported as a novel RA susceptibility gene locus in a meta-analysis of three GWASs for RA in the Japanese population, which included the GWAS set that the present study used [15]. RTKN2 is located in the same region (10q21) as ARID5B, in which a significant association signal was also reported in the meta-analysis [15]. In our GWAS set, however, two significant signals were observed at rs3125734 (RTKN2: P = 4.8×10−5) and rs10821944 (ARID5B: P = 7.4×10−4), the former of which was tested as a landmark in the replication study. These two SNPs were in weak LD (r2 = 0.11) and the independent effect of each SNP was observed after conditioning on each SNP (RTKN2: P = 1.5×10−3, ARID5B: P = 0.024, respectively). This indicated that two independent associations existed in this region, and the association of RTKN2 is novel. We also confirmed the association in the STAT4 locus [14] with genome-wide significance (2q32.2, rs10168266, P = 3.2×10−8, OR = 1.16, 95% CI = 1.10–1.22) (Table S2). The associations in NFKBIE and RTKN2 were further replicated in the 2nd replication cohort (3,417 cases and 3,763 controls; rs2233434, P = 1.1×10−5, OR = 1.19, 95% CI = 1.10–1.30 and rs3125734, P = 0.016, OR = 1.14, 95% CI = 1.02–1.26, respectively), confirming the associations in these loci (a combined analysis of three sets; rs2233434, P = 1.3×10−15, OR = 1.20, 95% CI = 1.15–1.26 and rs3125734, P = 4.6×10−9, OR = 1.20, 95% CI = 1.13–1.27, respectively) (Table 1 and Figure 1). We also genotyped these SNPs for individuals with systemic lupus erythematosus (SLE [MIM 152700]) (n = 657) and Graves' disease (GD [MIM 275000]) (n = 1,783). We identified a significant association of RTKN2 (rs3125734) with GD (P = 3.4×10−5, OR = 1.24, 95% CI = 1.12–1.37), whereas no significant associations were detected in NFKBIE (rs2233434) with either disease or in RTKN2 (rs3125734) with SLE (Table S3).
Fig. 1. Association plots of NFKBIE and RTKN2 regions. 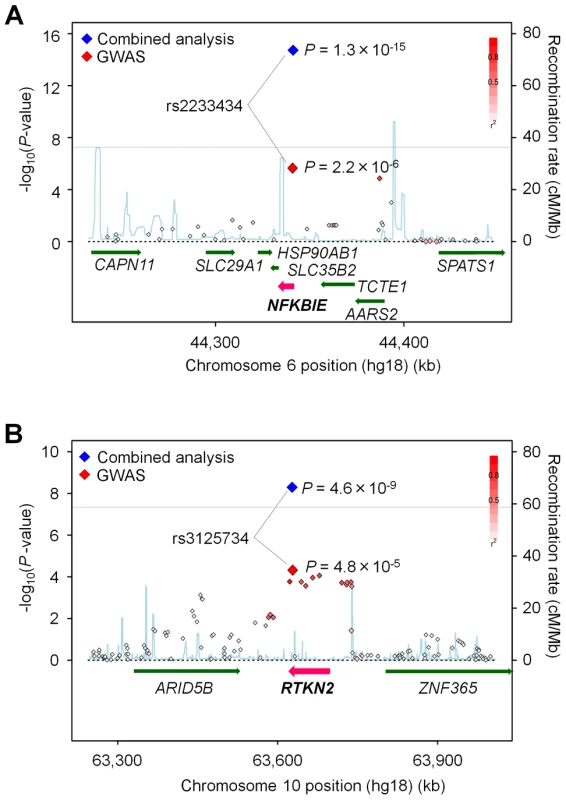
The diamonds represent the -log10 of the Cochran-Armitage trend P-values. Large diamonds show landmark SNPs in NFKBIE (rs2233434: A) and RTKN2 (rs3125734: B). Red: GWAS, Blue: combined analysis. Red colors of each SNP indicate its r2 with landmark SNP. Gray lines indicate the genome-wide significance threshold (P<5×10−8). For each plot, the -log10 P-values (y-axis) of the SNPs are presented according to their chromosomal positions (x-axis). Physical positions are based on NCBI build 36.3 of the human genome. Genetic recombination rates, estimated using the 1000 Genome Project (JPT and CHB), are represented by the blue line. Tab. 1. Association analysis of NFKBIE and RTKN2 with rheumatoid arthritis. 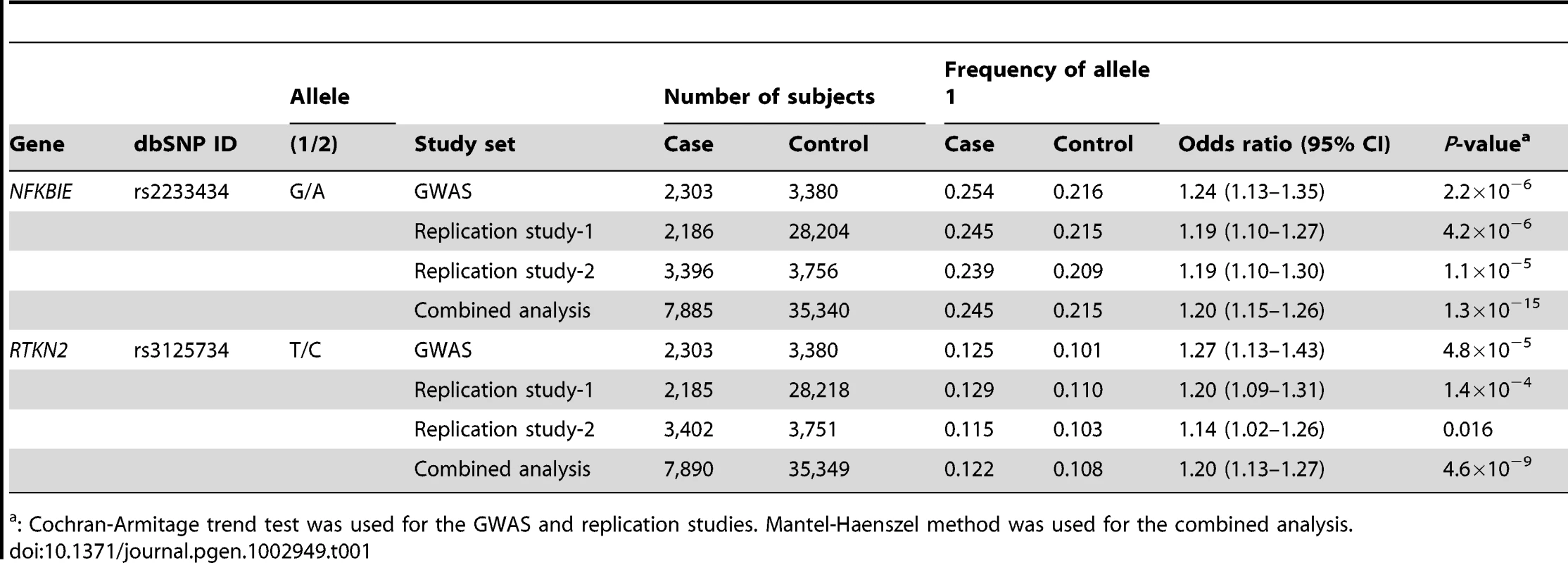
: Cochran-Armitage trend test was used for the GWAS and replication studies. Mantel-Haenszel method was used for the combined analysis. Functional analysis of non-synonymous SNPs
NFKBIE and RTKN2 genes are both involved in the NF-κB pathway: NFKBIE encodes IκB epsilon (IκBε), a member of the IκB family [16], and its binding to NF-κB inhibits the nuclear translocation of NF-κB [17]; RTKN2 encodes a member of Rho-GTPase effector proteins highly expressed in CD4+ T cells [18] and is implicated in the activation of the NF-κB pathway [19]. Considering that the NF-κB pathway is critical for the pathogenesis of RA [20], these two genes could be strong candidates in these regions. To identify disease-causing variants, we first sequenced the coding regions of the genes using DNA from patients (n = 48) to find variants that alter amino acid sequences. We identified four non-synonymous (ns)SNPs, which were all registered in the dbSNP database: two nsSNPs in NFKBIE (rs2233434 (Val194Ala) and rs2233433 (Pro175Leu)) and two in RTKN2 (rs3125734 (Arg462His) and rs61850830 (Ala288Thr)), where rs2233434 and rs3125734 were the same as the landmark SNPs in the GWAS (Figure 1 and Figure 2A). The two nsSNPs of each locus were in strong LD (Figure 2B) and were both associated with disease (Table S4). In the haplotype analysis, a single common risk haplotype with a frequency >0.05 was observed in each locus, and significant associations with disease risk were detected (NFKBIE, P = 5.3×10−8, Table S5; RTKN2, P = 5.7×10−5, Table S6).
Fig. 2. Genomic position and LD blocks. 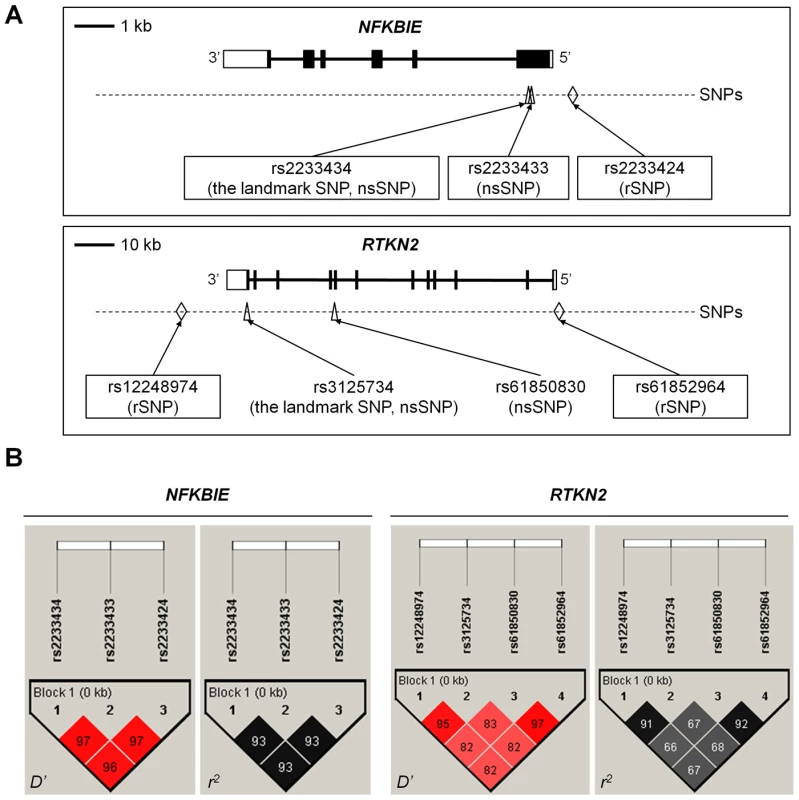
(A) Genomic position of non-synonymous (ns)SNPs and regulatory (r)SNPs in NFKBIE and RTKN2. NFKBIE (top) and RTKN2 (bottom) correspond to transcripts NM_004556.2 and NM_145307.2, respectively. Exons are shown as boxes, where black boxes represent coding regions and open boxes represent untranslated regions. Intron sequences are drawn as lines. Open triangles represents nsSNPs and open diamond shapes indicate candidate rSNPs. dbSNP IDs of candidate causal variants were boxed in a solid line. (B) LD patterns for nsSNPs and candidate rSNPs in NFKBIE (left) and RTKN2 (right) gene regions. LD blocks were constructed from genotype data of 3,290 control individuals of the GWAS. The diagrams show pairwise LD values as quantified using the D′ and r2 values. To investigate the effect of these nsSNPs on protein function, we evaluated them by in silico analysis using PolyPhen and SIFT software, which predicts possible impacts of amino acid substitutions on the structure and function of proteins, but all four nsSNPs were predicted to have little effect (Table S7), contrasting with the effect of Mendelian disease mutations [9]. We next examined their influence on the NF-κB activity in cells by performing NF-κB reporter assays with haplotype-specific expression vectors. In NFKBIE, the non-risk haplotype (A-C: rs2233434 (non-risk allele (NR))-rs2233433 (NR)) displayed an inhibitory effect on NF-κB activity compared with the mock construct, which reflected compulsorily binding of exogenous IκBε to the endogenous NF-κB, as shown in a previous study [16]. Of note, the risk haplotype (G-T: risk allele (R)-R) showed higher NF-κB activity than A-C (NR-NR) (Figure 3A), suggesting impaired inhibitory potential of G-T (R-R) products. No haplotypic difference was detected in the protein expression levels of these constructs (Figure 3C). We also examined two additional constructs of G-C (R-NR) and A-T (NR-R) haplotypes to evaluate the effect of each nsSNP (Figure S1A, S1B). Because NF-κB activity increased in the order of A-C<G-C<A-T<G-T (rs2233434-rs2233433: NR-NR<R-NR<NR-R<R-R) when cells were stimulated with TNF-α, the C>T substitution (Pro175Leu) in rs2233433 may have more impact on the protein function of IκBε compared with the A>G substitution (Val194Ala) in rs2233434. In contrast to the observations in NFKBIE, no clear difference was detected between the two common haplotype products of RTKN2 in either their effect on NF-κB activity or protein expression levels, although both products enhanced NF-κB activity as reported previously (Figure 3B, 3D) [19]. These functional analyses of nsSNPs suggest that two nsSNPs (rs2233434 and rs2233433) in the NFKBIE region are candidates for causal SNPs.
Fig. 3. Functional evaluation of nsSNPs and allelic imbalance of expression in NFKBIE and RTKN2. 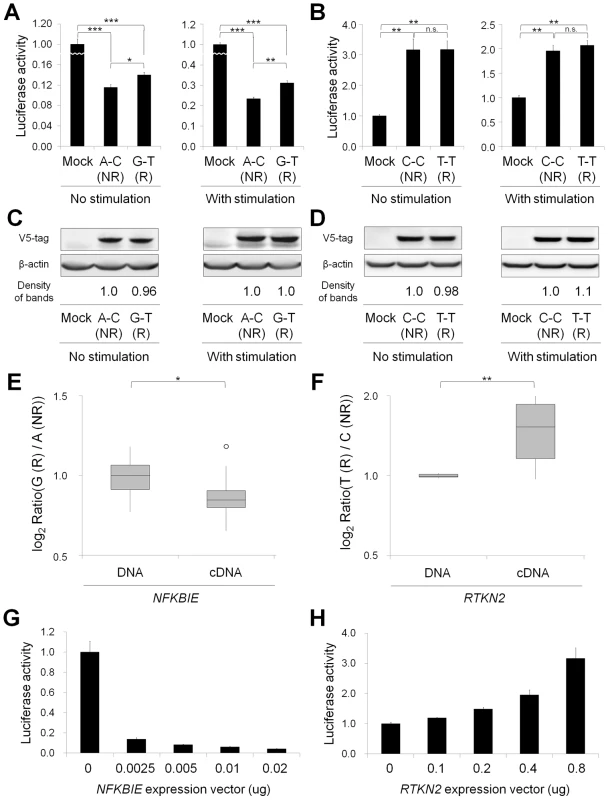
(A, B) Effects of nsSNPs in NFKBIE (A) and RTKN2 (B) on NF-κB activity by luciferase assays. Two haplotype constructs (A-C (rs2233434-rs2233433; non-risk (NR)) and G-T (risk (R)) for NFKBIE and C-C (rs3125734-rs61850830; NR) and T-T (R) for RTKN2) were used. The expression vector of each construct, pGL4.32[luc2P/NF-κB-RE] vector and pRL-TK vector were transfected into HEK293A cells. Data represent the mean ± s.d. Each experiment was performed in sextuplicate, and experiments were independently repeated three times. *P<0.05, **P<1.0×10−5, and ***P<1.0×10−10 by Student's t-test. n.s.: not significant. (C, D) Protein expression levels of each haplotype construct. Anti-V5 tag antibody was used in the Western blotting analysis to detect the expression of exogenous IκBε (C) and RTKN2 (D). Beta-actin expression was used as an internal control. The densities of the bands were quantified and normalized to that of the risk allele. (E, F) Allelic imbalance of expression in NFKBIE (E) and RTKN2 (F). ASTQ was performed using samples from individuals heterozygous for rs2233434 (G/A) in NFKBIE and rs3125734 (T/C) in RTKN2. Genomic DNAs and cDNAs were extracted from PBMCs (n = 14 for NFKBIE and n = 6 for RTKN2). The y-axis shows the log2 ratio of the transcript amounts in target SNPs (risk allele/non-risk allele). The top bar of the box-plot represents the maximum value and the lower bar represents the minimum value. The top of box is the third quartile, the bottom of box is the first quartile, and the middle bar is the median value. The circle is an outlier. *P = 0.012, **P = 0.016, by Student's t-test. (G, H) Dose-dependent inhibition of NFKBIE (G) and activation of RTKN2 (H) on NF-κB activity. Various doses of expression vectors carrying the non-risk allele of each gene were transfected into HEK293A cells with pGL4.32 and pRL-TK vectors. ASTQ analysis suggested the existence of regulatory variants
As the majority of autoimmune disease loci have been implicated as eQTL [11], we speculated that variants in the NFKBIE and RTKN2 loci would influence gene function by regulating gene expression, in addition to changing the amino acid sequences. To address this possibility, we performed allele-specific transcript quantification (ASTQ) analysis by using allele-specific probes targeting the nsSNPs in exons (rs2233434 for NFKBIE and rs3125734 for RTKN2, both of which were the GWAS landmarks). The genomic DNAs and cDNAs were extracted from peripheral blood mononuclear cells (PBMCs) in individuals with heterozygous genotype (n = 14 for NFKBIE and n = 6 for RTKN2) and from lymphoblastoid B-cell lines (n = 9) for NFKBIE. As the expression levels of RTKN2 were low in lymphoblastoid B cells, only PBMCs were used. When quantified by allele-specific probes, transcripts from the risk allele of NFKBIE showed 1.1-fold and 1.2-fold lower amounts (in PBMCs and lymphoblastoid B cells, respectively) than those from non-risk alleles (P = 0.012 and 5.3×10−4, respectively; Figure 3E and Figure S2). In contrast, 1.5-fold higher amounts of transcripts were observed in the risk allele of RTKN2 (P = 0.016; Figure 3F). These allelic imbalances suggested that both gene loci were eQTL and that there existed variants with cis-regulatory effects. Moreover, considering the inhibitory effects of NFKBIE and the activating potential of RTKN2 on NF-κB activity, which might both be dose dependent (Figure 3G, 3H), these regulatory variants in the risk alleles should enhance NF-κB activity in vivo.
Integrated in silico and in vitro analysis to search for regulatory variants
To comprehensively search the two genomic regions for causal regulatory variants, we performed an integrated in silico and in vitro analysis with multiple steps (Figure 4 and Figures S3, S4). We first determined the target genomic region by selecting LD blocks containing disease-associated SNPs (PGWAS<1.0×10−3) (Step 1). We then extracted SNPs with frequencies of >0.05 from HapMap and 1000 Genome Project databases in the region (Step 2). We excluded uncommon variants (MAF<0.05) from the analysis because of their low imputation accuracy in the GWAS (93% of uncommon variants in NFKBIE and 76% in RTKN2 exhibited Rsq <0.6). There is neither structural variation (>1 kbps) nor indels (100 bps to 1 kbs) that are common in the population (frequency >0.01) in these loci. To evaluate the cis-regulatory potential of sequences around the SNPs in silico, we used the regulatory potential (RP) score [21], [22]. This score was calculated based on the extent of sequence conservation among species or similarity with known regulatory motifs. We selected SNPs from the genomic elements with an RP score >0.1 (Step 3a). Subsequently, we selected SNPs from sites of transcriptional regulation as demonstrated by previous ChIP-seq studies (transcription factor binding sites [23], [24] and histone modification sites [25], [26]) or a DNase-seq study (DNase I hypersensitivity sites) [27] (Step 3b). Finally, these SNPs with regulatory potential were further screened out by the disease-association status (P<0.05) using an imputed GWAS dataset (Step 4). As a consequence, we selected 14 SNPs in NFKBIE and 10 SNPs in RTKN2 that had regulatory potential predicted in silico.
Fig. 4. Overview of SNP selection using integrated in silico and in vitro approaches. 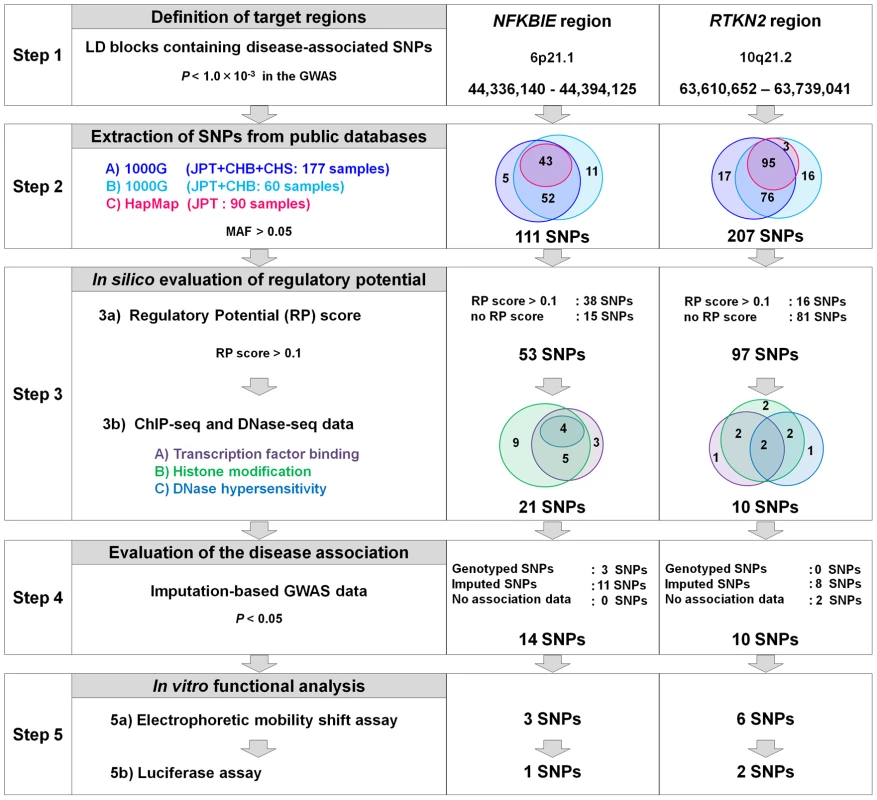
The figure shows the SNP selection process (left) and the results of NFKBIE (middle) and RTKN2 (right). (Step 1) LD blocks that contain disease-associated SNPs (PGWAS<1.0×10−3) were selected. (Step 2) SNPs were extracted from three databases (A–C). 1000G, 1000 Genome Project; HapMap, International HapMap Project. A) JPT, CHB, and CHS samples (n = 177) from the 1000G (the August 2010 release). B) JPT and CHB samples (n = 60) from the pilot 1 low coverage study data of 1000G (the March 2010 release). C) JPT samples (n = 90) from HapMap phase II+III (release #27). SNPs with minor allele frequency >0.05 were selected. (Step 3) Prediction of regulatory potential in silico. 3a) Regulatory potential (RP) scores were used for SNP selection, where an RP score >0.1 indicated the presence of regulatory elements. SNPs without RP scores were also selected. 3b) Prediction of regulatory elements by ChIP-seq data and DNase-seq data. (A) Transcription factor binding sites, (B) histone modification sites (CTCF binding, H3K4me1, H3K4me2, H3K4me3, H3K27ac, H3K9ac), and (C) DNase I hypersensitivity sites were evaluated. ChIP-seq and DNase-seq data derived from GM12878 EBV-transformed B cells were used for NFKBIE and RTKN2. DNase-seq data of Th1, Th2, and Jurkat cells were also used for RTKN2. (Step 4) Association data of the imputation-based GWAS using 1000G reference genotypes were used. SNPs with a significance level of P<0.05 were selected. SNPs without association data were also selected. (Step 5) EMSAs and luciferase assays were performed for evaluation of regulatory potentials in vitro. To further investigate the regulatory potential of the SNPs, we evaluated 31-bp sequences around the SNPs by in vitro assays. First, we examined their ability to bind nuclear proteins by EMSAs (Step 5a) using nuclear extracts from lymphoblastoid B cells (PSC cells) and Jurkat cells. Of the 24 SNPs examined, nine SNPs displayed allelic differences, implying differential potential of transcriptional activity between these alleles (Figure 5A and Figure S5). We then evaluated the enhancing or repressing activity of the sequences by luciferase reporter assays (Step 5b). We cloned them into the pGL4.24 vector, which has minimal promoter activity, and transfected these constructs into HEK293A cells (for NFKBIE and RTKN2), lymphoblastoid B cells (for NFKBIE), and Jurkat cells (for RTKN2). Among the three SNPs examined in NFKBIE, the risk allele of rs2233424 (located −396 bps from the 5′ end) displayed stronger repression activity (Figure 2A and Figure 5B) than that of the non-risk allele. Among the six SNPs in RTKN2, the risk alleles of rs12248974 (approximately 10 kb from the 3′ end) and rs61852964 (−215 bps from the 5′ end) showed higher enhancing activity compared with the non-risk alleles (Figure 2A and Figure 5B). These results corresponded to the results of ASTQ analyses (Figure 3E, 3F). Other SNPs showed no allelic differences or had the opposite trend of transcriptional activity in the risk allele compared to the results of ASTQ analysis (Figure S6).
Fig. 5. Evaluation of candidate regulatory SNPs in vitro. 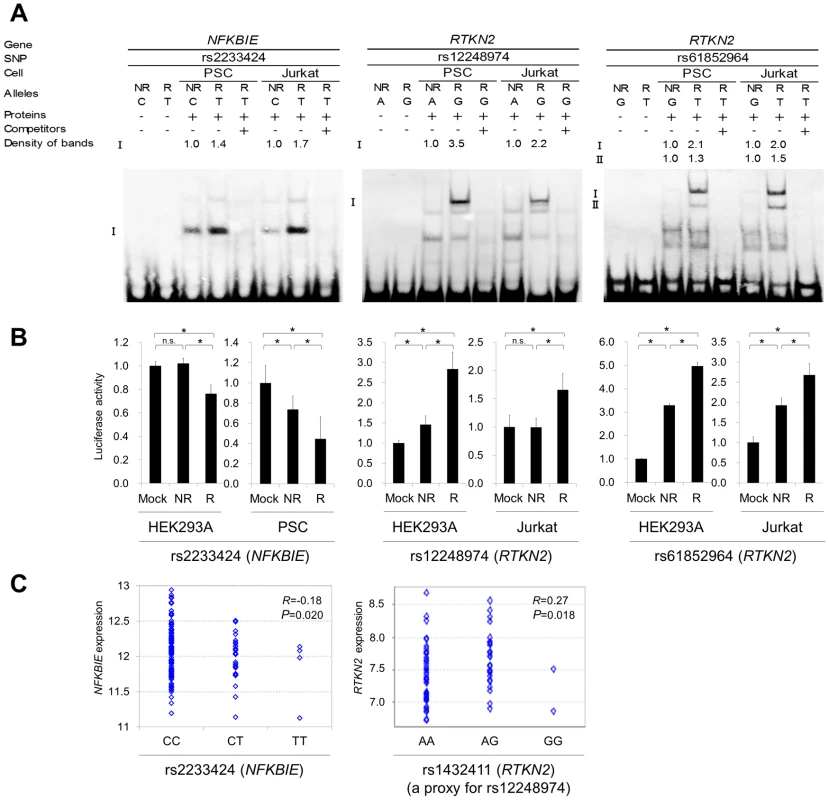
(A) Binding of nuclear factors from lymphoblastoid B-cells (PSC cells) and Jurkat cells to the 31-bp sequences around each SNP was evaluated by EMSA. Unlabeled probes in 200-fold excess as compared to the labeled probes were used for the competition experiment. The densities of the bands were quantified and normalized to that of the risk allele. rs2233424 in NFKBIE (C(NR)/T(R)) (left), rs12248974 (A(NR)/G(R)) (middle) and rs61852964 (G(NR)/T(R)) (right) in RTKN2. (B) Transcriptional activities were evaluated by luciferase assays. Each 31-bp oligonucleotide was inserted into the pGL4.24[Luc2P/minP] vector. Luc, luciferase; minP, minimal promoter. Transcfection was performed with HEK293A (for all the SNPs), PSC cells (for rs2233424), and Jurkat cells (for rs12248974 and rs61852964). rs2233424 (left), rs12248974 (middle), and rs61852964 (right). Data represent the mean ± s.d. Each experiment was performed in sextuplicate and independently repeated three times. *P<0.05 by Student's t-test. n.s.: not significant. (C) Liner regression analysis of the relationship between SNP genotype and gene expression level. NFKBIE expression data in lymphoblastoid B-cell lines of HapMap individuals (JPT+CHB, CEU and YRI; n = 151), and RTKN2 expression data in primary T cells from umbilical cords of Western European individuals (n = 85) were used. The x-axis shows the SNP genotypes and the y-axis represents the log2-transformed gene expression level. R: correlation coefficient between SNP genotype and gene expression. Rs2233424 genotypes and NFKBIE expression level (left). The genotype classification by population: JPT+CHB, CC = 52, CT = 1; CEU, CC = 35, CT = 2; YRI, CC = 32, CT = 2, TT = 4. Rs1432411 genotypes and RTKN2 expression level (right). Rs1432411 was used as a proxy SNP of rs12248974 (r2 = 0.97). To confirm the regulatory potential of these SNPs, we investigated the correlation between genotypes and gene expression levels in lymphocytes utilizing the data from the previous eQTL studies. We evaluated the expression of RTKN2 in primary T cells from Western European individuals by using Genevar software [28], [29]. Though NFKBIE is also expressed in primary T cells, the genotypes of rs2233424 are not available. We thus evaluated gene expression data of lymphoblastoid B-cell lines obtained from HapMap individuals (Japanese (JPT) + Han Chinese in Beijing (CHB), European (CEU), and African (YRI)) [30], [31] instead. The NFKBIE expression level decreased with the number of risk alleles of rs2233424 (R = −0.18, P = 0.020), and the RTKN2 expression levels increased with that of rs1432411 (a proxy for rs12248974, r2 = 0.97) (R = 0.27, P = 0.018) (Figure 5C), corresponding to the results of the in vitro assays. The data for rs61852964 in RTKN2 was not available. Among the SNPs that displayed opposite transcriptional activities in the reporter assays compared to the results of ASTQ, the data for rs2233434, rs77986492, and rs3852694 (a proxy for rs1864836, r2 = 1.0) were available (Figure S7 and S8). These SNPs displayed the opposite direction of the correlation trend as compared to the results of reporter assays, but parallel to ASTQ, implying that the regulatory effects observed in the in vitro assays were cancelled out by the effects of other regulatory variants on the same haplotype in vivo.
Finally, we validated the associations of these regulatory (r)SNPs observed in the imputed GWAS dataset. We directly genotyped them by TaqMan assay and confirmed significant associations (Table S8). As the candidate causal variants (nsSNPs and rSNPs) and the landmark SNPs of GWAS were in strong LD at each locus (Figure 2A, 2B), we evaluated the independent effect of each SNP by haplotype analysis in both loci (Table S9 and S10) and the conditional logistic regression analysis in RTKN2 (Table S11). The conditional analysis was not performed in NFKBIE because three candidate causal variants were in strong LD (r2>0.9). However, the analyses for these two loci did not demonstrate any evidence of primary or independent effects across the candidate causal variants, and it remains a possibility that all of the functional variants were involved in the pathogenesis. In addition, although the landmark nsSNP (rs3125734) in RTKN2 did not display any influence on NF-κB activity in our in vitro assays, rs3125734 might influence functions of RTKN2 other than those in the NF-κB pathway; alternatively, it is still possible that rs3125734 tags the effects of other unknown variants, such as rare variants, in addition to the other two rSNPs (rs12248974 and rs61852964).
Discussion
In the present study, we performed a replication study of our previously reported GWAS and identified variants in NFKBIE and RTKN2 loci that were associated with RA susceptibility. The associations of NFKBIE and RTKN2 loci have not been reported in other populations with genome-wide significance. However, rs2233434 in NFKBIE showed a suggestive association (589 cases vs. 1,472 controls, P = 0.0099, OR = 1.57, 95% CI = 1.11–2.21) in a previous meta-analysis in European populations [32]. The weak association signal in Europeans may be partially due to the lower frequency of the risk allele (0.04 in Europeans compared to 0.22 in Japanese). On the other hand, the association of rs3125734 in RTKN2 was not observed in a GWAS meta-analysis of European populations (cases 5,539 vs. controls 20,169, P = 0.11, OR = 1.04, 95% CI = 0.99–1.09). As the association of RTKN2 locus was also implicated in Graves' disease in a Han Chinese population [33], the association in RTKN2 locus may be unique to Asian populations.
To find the disease causal variants in disease-associated loci, target re-sequencing and variant genotyping with a large sample set followed by conditional association analysis examining the independent effects of each variant would be the first step. For this purpose, a recent attempt to fine-map the known autoimmunity risk loci in Celiac disease (MIM 212750) using an “Immunochip” brought us several insights [34]. First, no stronger signals compared to the GWAS signals were detected in most of the known loci, while additional independent signals were found in several loci. Second, none of the genome-wide significant common SNP signals could be explained by any rare highly penetrant variants. Third, although the fine-mapping strategy could localize the association signals into finer scale regions, it could not identify the actual causal variants due to strong LD among the variants, indicating that an additional approach, such as functional evaluation of candidate variants, is needed.
In the present study, we focused on common variants to find causal variants. Instead of re-sequencing additional samples, we utilized the 1000 Genome Project dataset, where the theoretically estimated cover rate for common variants (frequency of >0.05) in our population is >0.99 [12], [35]. To fine-map the association signals, we performed imputation-based association analysis, where we could not find any association signals that statistically exceeded the effect of landmark SNPs (rs2233434 for NFKBIE and rs3125734 for RTKN2) in both gene regions (Figures S3 and S4). We also performed a conditional logistic regression analysis, and found no additional independent signals of association when conditioned on each landmark SNP (data not shown). Although the imputation-based association tests may yield some bias compared to direct genotyping of the variants, these results suggested that variants in strong LD with the landmark SNPs were strong candidates for causal variants.
Following the analysis of nsSNPs, we evaluated cis-regulatory effects of variants in the two regions by ASTQ analysis using both B-cell lines and primary cells (PBMC), the majority of which consisted of T and B lymphocytes. As the mechanism of gene-regulation is substantially different between cell types [26], ASTQ analysis in more specific cell types that are relevant to the disease etiology, such as Th1 and Th17 cells, would be ideal to evaluate the cis-regulatory effects of variants. In this context, a more comprehensive catalog of the eQTL database of multiple cell types should be established for genetic study of diseases. As our ASTQ analysis demonstrated cis-regulatory effects of variants in both regions, we then performed an integrated in silico and in vitro analysis to identify candidate regulatory variants. Accumulating evidence by recent ChIP-seq and DNase-seq studies suggested that cis-regulatory variants are located in the key regions of transcriptional regulation [26], [36], warranting the prioritization of variants before evaluation by in vitro assays. This could also minimize false-positive results of the in vitro assays. However, there may be additional causal variants, including rare variants, unsuccessfully selected at each step of our integrated screening. Therefore, the screening strategy should be refined as the quality and quantity of genomic databases improves in the future.
We identified multiple candidate causal variants in NFKBIE (two nsSNPs and one rSNP) and RTKN2 (two rSNPs). We could not statistically distinguish the primary effect of each candidate causal variant, because these variants are in strong LD and on the same common haplotype. However, multiple causal variants could be involved in a single locus, which is also seen in another well-known autoimmune locus in 6q23 (TNFAIP3 gene locus), where both an nsSNP and a regulatory variant have been shown to be functionally related to the disease [8], [37]. The risk haplotype of nsSNPs in NFKBIE (rs2233433 and rs2233434) showed an enhancement of NF-κB activity, which might reflect an impaired inhibitory effect of IκB-ε on nuclear translocation of NF-κB. On the other hand, down-regulated NFKBIE expression and up-regulated RTKN2 expression were observed at the risk haplotypes, which may be regulated in cis by the rSNPs (rs2233424 in NFKBIE, rs12248974 and rs61852964 in RTKN2). As overexpression studies have also demonstrated dose-dependent attenuation of NF-κB activity by NFKBIE, and dose-dependent enhancement by RTKN2, the cis-regulatory effects of these rSNPs should enhance the NF-κB activity in the risk allele. Taken together with the effect of nsSNPs in NFKBIE, the enhancement of NF-κB activity may play a role in the pathogenesis of the disease. This is further supported by evidence that previous GWAS for RA have also identified genes related to the NF-κB pathway, such as TNFAIP3 [13], v-rel reticuloendotheliosis viral oncogene homolog (REL [MIM 164910]) [5], TNF receptor-associated factor 1 (TRAF1 [MIM 601711]) [3], and CD40 molecule TNF receptor superfamily member 5 (CD40 [MIM 109535]) [38].
In conclusion, we identified NFKBIE and RTKN2 as genetic risk factors for RA. Considering the allelic effect of both genes, enhanced NF-κB activity may play a role in the pathogenesis of the disease. Because NF-κB regulates the expression of numerous genes, including inflammatory and immune response mediators, NF-κB and its regulators identified by GWAS are promising targets for the treatment of RA.
Materials and Methods
Ethics statement
All subjects were of Japanese origin and provided written informed consent for participation in the study, which was approved by the ethical committees of the institutional review boards.
Subjects
A total of 7,907 RA cases, 657 SLE cases, 1,783 GD cases, and 35,362 control subjects were enrolled in the study through medical institutes in Japan under the support of the BioBank Japan Project, Center for Genomic Medicine at RIKEN, the University of Tokyo, Tokyo Women's Medical University, and Kyoto University. The same case and control samples were used in the previous meta-analysis of GWASs in the Japanese population (Table S1) [15] . RA and SLE subjects met the revised American College of Rheumatology (ACR) criteria for RA [39]. Diagnosis of individuals with GD was established on the basis of clinical findings and results of the routine examinations for circulating thyroid hormone and thyroid-stimulating hormone concentrations, thyroid-stimulating hormone receptors, ultrasonography, [99m]TCO4− (or [123I]) uptake, and thyroid scintigraphy. DNAs were extracted from peripheral blood cells using a standard protocol. Total RNAs were also extracted from PBMCs of healthy individuals (n = 20) using an RNeasy kit (QIAGEN, Valencia, CA, USA). Details of the samples are summarized in Table S1.
Genotyping and quality control
In the GWAS, RA cases and controls were genotyped using Illumina Human610-Quad and Illumina Human 550v3 Genotyping BeadsChips (Illumina, San Diego, CA, USA), respectively, and quality control of genotyping was performed as described previously [6]. For replication study of candidate loci, a landmark SNP was selected from each locus that satisfied 5×10−8<PGWAS<5×10−5 in the GWAS. If multiple candidate SNPs existed within ±100 kb, the SNP with the lowest P-value was selected. All case subjects in the replication study and both case and control subjects in the validation study of candidate causal variants were genotyped using TaqMan SNP genotyping assays (Table S12) (Applied Biosystems, Foster City, CA, USA) with an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Because of the availability of DNA samples, only a part of the control subjects were genotyped for the validation study (n = 3,290, 97.3%). To enlarge the number of subjects and enhance statistical power for replication studies, we used genotype data obtained from other GWAS projects genotyped using the Illumina platforms for the replication control panels (Table S1). All SNPs were successfully genotyped with call rates >0.98 and were in Hardy-Weinberg equilibrium (HWE) in control subjects (P>0.05 as examined by χ2 test), except for rs2233434, which displayed a deviation from HWE (P = 0.00091). To evaluate possible genotyping biases between the platforms, we also genotyped rs2233434 and rs3125734 by TaqMan assays for randomly selected subjects genotyped using other genotyping platforms (n = 376), yielding high concordance rates of ≥0.99.
Association analysis
The associations of the SNPs were tested with the Cochran-Armitage trend test. Combined analysis was performed with the Mantel-Haenszel method. Haplotype association analysis and haplotype-based conditional association analysis were performed using Haploview v4.2 and the PLINK v1.07 program (see URLs) [40], respectively. The SNPs that were not genotyped in the GWAS were imputed using MACH 1.0.16 (see URLs), with genotype data from the 1000 Genome Project (JPT, CHB, and Han Chinese South (CHS): 177 individuals) as references (August 2010 release) [41]. All the imputed SNPs demonstrated Rsq values more than 0.60.
DNA re-sequencing
Unknown variants in the coding sequences of NFKBIE and RTKN2 were revealed by directly sequencing the DNA of 48 individuals affected with RA. DNA fragments were amplified with the appropriate primers (Table S13). Purification of PCR products was performed with Exonuclease I (New England Biolabs, Ipswich, MA, USA) and shrimp alkaline phosphatase (Promega, Madison, WI, USA). The amplified DNAs were sequenced using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems), and signals were detected using an ABI 3700 DNA Analyzer (Applied Biosystems).
Construction of haplotype-specific expression vectors
The full coding regions were amplified using cDNAs prepared from an Epstein-Barr virus-transfected lymphoblastoid B-cell line (Pharma SNP Consortium (PSC), Osaka, Japan) for NFKBIE (NM_004556.2) and from Jurkat cells (American Type Culture Collection (ATCC), Rockville, MD, USA) for RTKN2 (NM_145307.2) with appropriate primers (Table S14) and DNA polymerases. PCR products were inserted into the pcDNA3.1D/V5-His-TOPO vector (Invitrogen, Camarillo, CA, USA) using the TaKaRa Ligation kit ver. 2.1 (Takara Bio Inc, Shiga, Japan), and mutagenized using the AMAP Multi Site-Directed Mutagenesis Kit (MBL, Nagoya, Japan). Each construct was then transformed into Jet Competent Escherichia coli cells (DH5α) (BioDynamics Laboratory Inc., Tokyo, Japan). These plasmids were purified using an Endofree Plasmid Maxi Kit (QIAGEN) after confirmation of the sequence.
NF-κB reporter assay
Human embryonic kidney (HEK) 293A cells (Invitrogen) were cultured in Dulbecco's modified Eagle's medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (BioWest, Nuaillé, France), 1% penicillin/streptomycin (Invitrogen), and 0.1 mM MEM Non-Essential Amino Acids (Invitrogen). Various doses of the haplotype-specific expression vector (0.0025–0.02 µg for NFKBIE and 0.1–0.8 µg for RTKN2), pGL4.32[luc2P/NF-κB-RE/Hygro] vector (Promega) (0.05 µg and 0.0125 µg, respectively), and pRL-TK vector (an internal control for transfection efficiency) (0.45 µg and 0.15 µg, respectively) were transfected into the HEK293A cells using the Lipofectamine LTX transfection reagent (Invitrogen) according to the manufacturer's protocol. The total amounts of DNAs were adjusted with empty pcDNA3.1 vector. After 22 h, cells were incubated with 1 ng/ml TNF-α (Sigma) for 2 h or with medium alone. Cells were collected, and luciferase activity was measured using a Dual-Luciferase Reporter Assay system (Promega) and a GloMax-Multi+ Detection System (Promega). Each experiment was independently repeated three times, and sextuplicate samples were assayed each time.
Western blotting
After 24 h of transfection as described for the NF-κB reporter assay, cells were lysed in NP-40 lysis buffer (150 mM NaCl, 1% NP-40, 50 mM Tris-HCl at pH 8.0, and a protease inhibitor cocktail), and incubated on ice for 30 min. After centrifugation, the supernatant fraction was collected and 4×Sodium dodecyl sulfate (SDS) sample buffer was added. After denaturation at 95°C for 5 min, proteins were analyzed by SDS-polyachrylamide gel electrophoresis (PAGE) on a 5% to 20% gradient gel (Wako, Osaka, Japan) and were transferred to polyvinylidene difluouride (PVDF) membranes (Millipore, Billerica, MA, USA). Target proteins on the membrane were probed with antibodies (mouse anti-V5 tag (Invitrogen), anti-β-actin-HRP (an internal control), and goat anti-mouse IgG2a-HRP (Santa Cruz Biotechnology, Santa Cruz, CA, USA)), visualized using enhanced chemiluminescence (ECL) detection reagent (GE Healthcare, Pollards Wood, UK), and detected using a LAS-3000 mini lumino-image analyzer (Fujifilm, Tokyo, Japan). Band intensities were measured using MultiGauge software (Fujifilm).
Allele-specific transcript quantification (ASTQ) analysis
ASTQ analysis was performed as previously described [42]. Total RNAs and genomic DNAs were extracted from PBMCs and lymphoblastoid B-cell lines. cDNAs were synthesized using TaqMan reverse transcription reagents (Applied Biosystems). We selected SNPs (rs2233434 (A/G) for NFKBIE and rs3125734 (C/T) for RTKN2) as target SNPs. Allele-specific gene expression was measured by TaqMan SNP genotyping probes for these SNPs (Applied Biosystems). To make a standard curve, we selected two individuals that had homozygous genotypes of each target SNP. We mixed these DNAs at nine different ratios and detected the intensities. The log2 of (risk allele/non-risk allele intensity) for each SNP was plotted against the log2 of mixing homozygous DNAs. We generated a standard curve (linear regression line; y = ax+b), where y is the log2 of (risk allele/non-risk allele intensity) at a given mixing ratio, x is the log2 of the mixing ratio, a is the slope, and b is the intercept. We then measured the allelic ratio for each cDNA and genomic DNA from each individual by real-time TaqMan PCR. Based on a standard curve, we calculated the allelic ratio of cDNAs and genomic DNAs. Intensities were detected using an ABI Prism 7900HT Sequence Detection System (Applied Biosystems).
Electrophoretic mobility shift assays (EMSA)
EMSA and preparation of nuclear extract from lymphoblastoid B-cell lines and Jurkat cells were performed as previously described [43]. Cells were cultured in RPMI-1640 medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Following stimulation with 50 ng/ml phorbol myristate acetate (Sigma-Aldrich) for 2 h, cells were collected and suspended in buffer A (20 mM HEPES at pH 7.6, 20% glycerol, 10 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA at pH 8.0, 1 mM DTT, 0.1% NP-40, and a protease inhibitor cocktail) for 10 min on ice. After centrifugation, the pellets were resuspended in buffer B (which contains buffer A with 500 mM NaCl). Following incubation on ice for 30 min and centrifugation to remove cellular debris, the supernatant fraction containing nuclear proteins was collected. Oligonucleotides (31-bp) were designed that corresponded to genomic sequences surrounding the SNPs (Table S15). Single-stranded oligonucleotide probes were labeled using a Biotin 3′ End DNA Labeling Kit (Pierce Biotechnology, Rockford, IL, USA), and sense and antisense oligonucleotides were then annealed. DNA-protein interactions were detected using a LightShift Chemiluminescent EMSA kit (Pierce Biotechnology). The DNA-protein complexes were separated on a non-denaturing 5% polyachrylamide gel in 1×TBE (Tris-borate-EDTA) running buffer for 60 min at 150 V. The DNA-protein complexes were then transferred from the gel onto a nitrocellulose membrane (Ambion, Carlsbad, CA, USA), and were cross-linked to the membrane by exposure to UV light. Signals were detected using a LAS-3000 mini lumino-image analyzer (Fujifilm). Allelic differences were analyzed using MultiGauge software (Fujifilm) by measuring the intensity of the bands.
Luciferase assay
Oligonucleotides (31-bp) were designed as described for the EMSAs (Table S15), and complementary sense and antisense oligonucleotides were annealed. To construct luciferase reporter plasmids, pGL4.24[luc2P/minP] vector (Promega) was digested with restriction enzymes (XhoI and BglII) (Takara Bio Inc), and annealed oligonucleotide was ligated into a pGL4.24 vector upstream of the minimal promoter. HEK293A (n = 2.5×105), lymphoblastoid B-cell lines (n = 2.0×106) and Jurkat (n = 5.0×105) cells were transfected with the allele-specific constructs (0.4 µg, 1.8 µg and 2.5 µg, respectively) and the pRL-TK vector (0.1 µg, 0.2 µg and 0.25 µg, respectively) using the Lipofectamine LTX transfection reagent (for HEK293A and Jurkat cells) and Amaxa nucleofector kit (Lonza, Basel, Switzerland) (for lymphoblastoid B-cell lines). Cells were collected, and luciferase activity was measured as described for the NF-κB reporter assay. Each experiment was independently repeated three times and sextuplicate samples were assayed each time.
Correlation analysis between gene expression and genotypes
The expression data in lymphoblastoid B-cell lines derived from HapMap individuals (n = 210; JPT, CHB, CEU, and YRI) and in primary T cells from umbilical cords of Western European individuals (n = 85) from the database of the Gene Expression Variation (Genevar) project were used. SNP genotypes were obtained from HapMap and 1000 Genome Project databases. The expression levels were regressed with the genotype in a liner model. The statistical significance of regression coefficients was tested using Student's t-test.
Statistical analysis
We used χ2 contingency table tests to evaluate the significance of differences in allele frequency in the case-control subjects. We defined haplotype blocks using the solid spine of LD definition of Haploview v4.2, and estimated haplotype frequency and calculated pairwise LD indices (r2) between pairs of polymorphisms using the Haploview program. Luciferase assay data and ASTQ analysis data were analyzed by Student's t-test.
Web resources
The URLs for data presented herein are as follows:
PLINK, http://pngu.mgh.harvard.edu/~purcekk/plink
MACH, http://www.sph.umich.edu/csg/abecasis/mach/
UCSC Genome Browser, http://genome.ucsc.edu/;
Genevar, http://www.sanger.ac.uk/resources/software/genevar/
HapMap Project, http://www.HapMap.org/
1000 Genome Project, http://www.1000genomes.org
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
Supporting Information
Zdroje
1. GabrielSE (2001) The epidemiology of rheumatoid arthritis. Rheum Dis Clin North Am 27 : 269–281.
2. SuzukiA, YamadaR, ChangX, TokuhiroS, SawadaT, et al. (2003) Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet 34 : 395–402.
3. PlengeRM, SeielstadM, PadyukovL, LeeAT, RemmersEF, et al. (2007) TRAF1-C5 as a risk locus for rheumatoid arthritis–a genomewide study. N Engl J Med 357 : 1199–1209.
4. Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447 : 661–678.
5. GregersenPK, AmosCI, LeeAT, LuY, RemmersEF, et al. (2009) REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet 41 : 820–823.
6. KochiY, OkadaY, SuzukiA, IkariK, TeraoC, et al. (2010) A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat Genet 42 : 515–519.
7. BegovichAB, CarltonVE, HonigbergLA, SchrodiSJ, ChokkalingamAP, et al. (2004) A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet 75 : 330–337.
8. AdriantoI, WenF, TempletonA, WileyG, KingJB, et al. (2011) Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat Genet 43 : 253–258.
9. ThomasPD, KejariwalA (2004) Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: evolutionary evidence for differences in molecular effects. Proc Natl Acad Sci U S A 101 : 15398–15403.
10. OkadaY, ShimaneK, KochiY, TahiraT, SuzukiA, et al. (2012) A Genome-Wide Association Study Identified AFF1 as a Susceptibility Locus for Systemic Lupus Eyrthematosus in Japanese. PLoS Genet 8: e1002455 doi:10.1371/journal.pgen.1002455.
11. DuboisPC, TrynkaG, FrankeL, HuntKA, RomanosJ, et al. (2010) Multiple common variants for celiac disease influencing immune gene expression. Nat Genet 42 : 295–302.
12. 1000 Genomes Project Consortium (2010) A map of human genome variation from population-scale sequencing. Nature 467 : 1061–1073.
13. PlengeRM, CotsapasC, DaviesL, PriceAL, de BakkerPI, et al. (2007) Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet 39 : 1477–1482.
14. RemmersEF, PlengeRM, LeeAT, GrahamRR, HomG, et al. (2007) STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med 357 : 977–986.
15. OkadaY, TeraoC, IkariK, KochiY, OhmuraK, et al. (2012) Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet 45 : 511–516.
16. LiZ, NabelGJ (1997) A new member of the I kappaB protein family, I kappaB epsilon, inhibits RelA (p65)-mediated NF-kappaB transcription. Mol Cell Biol 17 : 6184–6190.
17. WhitesideST, EpinatJC, RiceNR, IsraelA (1997) I kappa B epsilon, a novel member of the I kappa B family, controls RelA and cRel NF-kappa B activity. Embo J 16 : 1413–1426.
18. CollierFM, Gregorio-KingCC, GoughTJ, TalbotCD, WalderK, et al. (2004) Identification and characterization of a lymphocytic Rho-GTPase effector: rhotekin-2. Biochem Biophys Res Commun 324 : 1360–1369.
19. CollierFM, LovingA, BakerAJ, McLeodJ, WalderK, et al. (2009) RTKN2 Induces NF-kappaB Dependent Resistance to Intrinsic Apoptosis in HEK cells and Regulates BCL-2 Gene in Human CD4+ Lymphocytes. J Cell Death 2 : 9–23.
20. MakarovSS (2001) NF-kappa B in rheumatoid arthritis: a pivotal regulator of inflammation, hyperplasia, and tissue destruction. Arthritis Res 3 : 200–206.
21. KolbeD, TaylorJ, ElnitskiL, EswaraP, LiJ, et al. (2004) Regulatory potential scores from genome-wide three-way alignments of human, mouse, and rat. Genome Res 14 : 700–707.
22. TaylorJ, TyekuchevaS, KingDC, HardisonRC, MillerW, et al. (2006) ESPERR: learning strong and weak signals in genomic sequence alignments to identify functional elements. Genome Res 16 : 1596–1604.
23. JohnsonDS, MortazaviA, MyersRM, WoldB (2007) Genome-wide mapping of in vivo protein-DNA interactions. Science 316 : 1497–1502.
24. ValouevA, JohnsonDS, SundquistA, MedinaC, AntonE, et al. (2008) Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods 5 : 829–834.
25. MikkelsenTS, KuM, JaffeDB, IssacB, LiebermanE, et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448 : 553–560.
26. ErnstJ, KheradpourP, MikkelsenTS, ShoreshN, WardLD, et al. (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473 : 43–49.
27. SaboPJ, KuehnMS, ThurmanR, JohnsonBE, JohnsonEM, et al. (2006) Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods 3 : 511–518.
28. DimasAS, DeutschS, StrangerBE, MontgomerySB, BorelC, et al. (2009) Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325 : 1246–1250.
29. YangTP, BeazleyC, MontgomerySB, DimasAS, Gutierrez-ArcelusM, et al. (2010) Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics 26 : 2474–2476.
30. StrangerBE, ForrestMS, DunningM, IngleCE, BeazleyC, et al. (2007) Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315 : 848–853.
31. StrangerBE, NicaAC, ForrestMS, DimasA, BirdCP, et al. (2007) Population genomics of human gene expression. Nat Genet 39 : 1217–1224.
32. StahlEA, RaychaudhuriS, RemmersEF, XieG, EyreS, et al. (2010) Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet 42 : 508–514.
33. ChuX, PanCM, ZhaoSX, LiangJ, GaoGQ, et al. (2011) A genome-wide association study identifies two new risk loci for Graves' disease. Nat Genet 43 : 897–901.
34. TrynkaG, HuntKA, BockettNA, RomanosJ, MistryV, et al. (2011) Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet 43 : 1193–1201.
35. LiY, SidoreC, KangHM, BoehnkeM, AbecasisGR (2011) Low-coverage sequencing: implications for design of complex trait association studies. Genome Res 21 : 940–951.
36. DegnerJF, PaiAA, Pique-RegiR, VeyrierasJB, GaffneyDJ, et al. (2012) DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482 : 390–394.
37. MusoneSL, TaylorKE, LuTT, NitithamJ, FerreiraRC, et al. (2008) Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet 40 : 1062–1064.
38. RaychaudhuriS, RemmersEF, LeeAT, HackettR, GuiducciC, et al. (2008) Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet 40 : 1216–1223.
39. ArnettFC, EdworthySM, BlochDA, McShaneDJ, FriesJF, et al. (1988) The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 31 : 315–324.
40. PurcellS, NealeB, Todd-BrownK, ThomasL, FerreiraMA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 : 559–575.
41. LiY, WillerC, SannaS, AbecasisG (2009) Genotype imputation. Annu Rev Genomics Hum Genet 10 : 387–406.
42. AkamatsuS, TakataR, AshikawaK, HosonoN, KamataniN, et al. (2010) A functional variant in NKX3.1 associated with prostate cancer susceptibility down-regulates NKX3.1 expression. Hum Mol Genet 19 : 4265–4272.
43. AndrewsNC, FallerDV (1991) A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res 19 : 2499.
Štítky
Genetika Reprodukční medicína
Článek Genome-Wide Association Study for Serum Complement C3 and C4 Levels in Healthy Chinese SubjectsČlánek Role of Transposon-Derived Small RNAs in the Interplay between Genomes and Parasitic DNA in RiceČlánek Tetraspanin Is Required for Generation of Reactive Oxygen Species by the Dual Oxidase System inČlánek An Essential Role of Variant Histone H3.3 for Ectomesenchyme Potential of the Cranial Neural CrestČlánek A Mimicking-of-DNA-Methylation-Patterns Pipeline for Overcoming the Restriction Barrier of BacteriaČlánek A Genome-Wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 9
-
Všechny články tohoto čísla
- Heterozygous Mutations in DNA Repair Genes and Hereditary Breast Cancer: A Question of Power
- GWAS of Diabetic Nephropathy: Is the GENIE out of the Bottle?
- The Conflict within and the Escalating War between the Sex Chromosomes
- Proteome-Wide Analysis of Disease-Associated SNPs That Show Allele-Specific Transcription Factor Binding
- Exome Sequencing Identifies Rare Deleterious Mutations in DNA Repair Genes and as Potential Breast Cancer Susceptibility Alleles
- A Gene Family Derived from Transposable Elements during Early Angiosperm Evolution Has Reproductive Fitness Benefits in
- Genome-Wide Association Study for Serum Complement C3 and C4 Levels in Healthy Chinese Subjects
- Role of Transposon-Derived Small RNAs in the Interplay between Genomes and Parasitic DNA in Rice
- Co-Evolution of Mitochondrial tRNA Import and Codon Usage Determines Translational Efficiency in the Green Alga
- SIRT6/7 Homolog SIR-2.4 Promotes DAF-16 Relocalization and Function during Stress
- CNV Formation in Mouse Embryonic Stem Cells Occurs in the Absence of Xrcc4-Dependent Nonhomologous End Joining
- Tetraspanin Is Required for Generation of Reactive Oxygen Species by the Dual Oxidase System in
- Citrullination of Histone H3 Interferes with HP1-Mediated Transcriptional Repression
- Variation in Genes Related to Cochlear Biology Is Strongly Associated with Adult-Onset Deafness in Border Collies
- The Long Non-Coding RNA Affects Chromatin Conformation and Expression of , but Does Not Regulate Its Imprinting in the Developing Heart
- Rif2 Promotes a Telomere Fold-Back Structure through Rpd3L Recruitment in Budding Yeast
- Is a Metastasis Susceptibility Gene That Suppresses Metastasis by Modifying Tumor Interaction with the Cell-Mediated Immunity
- The p38/MK2-Driven Exchange between Tristetraprolin and HuR Regulates AU–Rich Element–Dependent Translation
- Rare Copy Number Variants Contribute to Congenital Left-Sided Heart Disease
- A Genetic Basis for a Postmeiotic X Versus Y Chromosome Intragenomic Conflict in the Mouse
- An Essential Role of Variant Histone H3.3 for Ectomesenchyme Potential of the Cranial Neural Crest
- Characterization of Inducible Models of Tay-Sachs and Related Disease
- Hominoid-Specific Protein-Coding Genes Originating from Long Non-Coding RNAs
- Transcriptional Repression of Hox Genes by HP1/HPL and H1/HIS-24
- Integrative Genomic Analysis Identifies Isoleucine and CodY as Regulators of Virulence
- Convergence of the Transcriptional Responses to Heat Shock and Singlet Oxygen Stresses
- Genomics of Adaptation during Experimental Evolution of the Opportunistic Pathogen
- Enrichment of HP1a on Drosophila Chromosome 4 Genes Creates an Alternate Chromatin Structure Critical for Regulation in this Heterochromatic Domain
- Vsx2 Controls Eye Organogenesis and Retinal Progenitor Identity Via Homeodomain and Non-Homeodomain Residues Required for High Affinity DNA Binding
- The Long Path from QTL to Gene
- TCF7L2 Modulates Glucose Homeostasis by Regulating CREB- and FoxO1-Dependent Transcriptional Pathway in the Liver
- The Non-Flagellar Type III Secretion System Evolved from the Bacterial Flagellum and Diversified into Host-Cell Adapted Systems
- Complex Chromosomal Rearrangements Mediated by Break-Induced Replication Involve Structure-Selective Endonucleases
- Factors That Promote H3 Chromatin Integrity during Transcription Prevent Promiscuous Deposition of CENP-A in Fission Yeast
- A Mimicking-of-DNA-Methylation-Patterns Pipeline for Overcoming the Restriction Barrier of Bacteria
- Determinants of Human Adipose Tissue Gene Expression: Impact of Diet, Sex, Metabolic Status, and Genetic Regulation
- Genome-Wide Association Studies Identify Heavy Metal ATPase3 as the Primary Determinant of Natural Variation in Leaf Cadmium in
- Tethering of the Conserved piggyBac Transposase Fusion Protein CSB-PGBD3 to Chromosomal AP-1 Proteins Regulates Expression of Nearby Genes in Humans
- A Genome-Wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans
- Normal DNA Methylation Dynamics in DICER1-Deficient Mouse Embryonic Stem Cells
- H4K20me1 Contributes to Downregulation of X-Linked Genes for Dosage Compensation
- The NDR Kinase Scaffold HYM1/MO25 Is Essential for MAK2 MAP Kinase Signaling in
- Coevolution within and between Regulatory Loci Can Preserve Promoter Function Despite Evolutionary Rate Acceleration
- New Susceptibility Loci Associated with Kidney Disease in Type 1 Diabetes
- SWI/SNF-Like Chromatin Remodeling Factor Fun30 Supports Point Centromere Function in
- A Response Regulator Interfaces between the Frz Chemosensory System and the MglA/MglB GTPase/GAP Module to Regulate Polarity in
- Functional Variants in and Involved in Activation of the NF-κB Pathway Are Associated with Rheumatoid Arthritis in Japanese
- Two Distinct Repressive Mechanisms for Histone 3 Lysine 4 Methylation through Promoting 3′-End Antisense Transcription
- Genetic Modifiers of Chromatin Acetylation Antagonize the Reprogramming of Epi-Polymorphisms
- UTX and UTY Demonstrate Histone Demethylase-Independent Function in Mouse Embryonic Development
- A Comparison of Brain Gene Expression Levels in Domesticated and Wild Animals
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Enrichment of HP1a on Drosophila Chromosome 4 Genes Creates an Alternate Chromatin Structure Critical for Regulation in this Heterochromatic Domain
- Normal DNA Methylation Dynamics in DICER1-Deficient Mouse Embryonic Stem Cells
- The NDR Kinase Scaffold HYM1/MO25 Is Essential for MAK2 MAP Kinase Signaling in
- Functional Variants in and Involved in Activation of the NF-κB Pathway Are Associated with Rheumatoid Arthritis in Japanese
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



