-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaFocusing In on the Complex Genetics of Myopia
article has not abstract
Published in the journal: . PLoS Genet 9(4): e32767. doi:10.1371/journal.pgen.1003442
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1003442Summary
article has not abstract
Refractive Errors Are Important Public Health Problems
Myopia (i.e., nearsightedness) is rapidly becoming a significant public health problem. The incidence of myopia has been increasing for at least three decades [1], [2], and 2.5 billion people will be affected by myopia by the year 2020 [3]. In some East Asian countries, the prevalence of myopia now exceeds 70% among teenagers and young adults [2], [4], [5]. Even though ocular refraction (the quantitative trait underlying myopia) is highly heritable [6], the recent change in the incidence of myopia is obviously not the result of short-term shifts in the genetic makeup of the population. Instead, secular trends in environmental and behavioral factors are thought to be driving the myopia “epidemic” throughout the world.
The Etiology of Myopia: A Historical Dilemma
The etiology of myopia has been the subject of conjecture for centuries. Johannes Kepler, who published the first comprehensive treatise on the optics of the eye and myopia in 1604 [7], attributed his own nearsightedness to intense study. Ever since, scientists have engaged in a heated nature versus nurture debate on the causes of myopia. Inherited and congenital forms of myopia have been documented since at least 1906 [8], [9]. Not until the late 1970s did experiments conclusively show that myopia could be induced during postnatal development by altering the visual experience of animal models through form deprivation or optical defocus [10]–[12]. Subsequent human and animal studies have led to the development of a robust model for refractive control during infancy, dubbed the emmetropization process. Although the details of this mechanism are still emerging, its effect is to maintain homeostasis by adjusting eye growth so that the plane of the retina coincides with the focal point of the eye's optical system, much like a camera lens focuses an image on its sensor. In this paradigm, altered visual stimuli (e.g., optically blurred images) initiate a signaling cascade that originates locally in the sensory retina, traverses the retinal pigment epithelium and the vascular choroid, and ultimately regulates eye growth via active remodeling of the sclera—the rigid white connective tissue that forms the outer layer of the eye globe (Figure 1, panels A and B). Alterations within this sequence of biochemical events may disrupt this finely tuned mechanism and lead to refractive errors, resulting in blurred vision. Hence, any gene that plays a role in this complex signaling pathway may contain susceptibility variants for myopia.
Fig. 1. The physical, biological, and genetic basis of myopia. 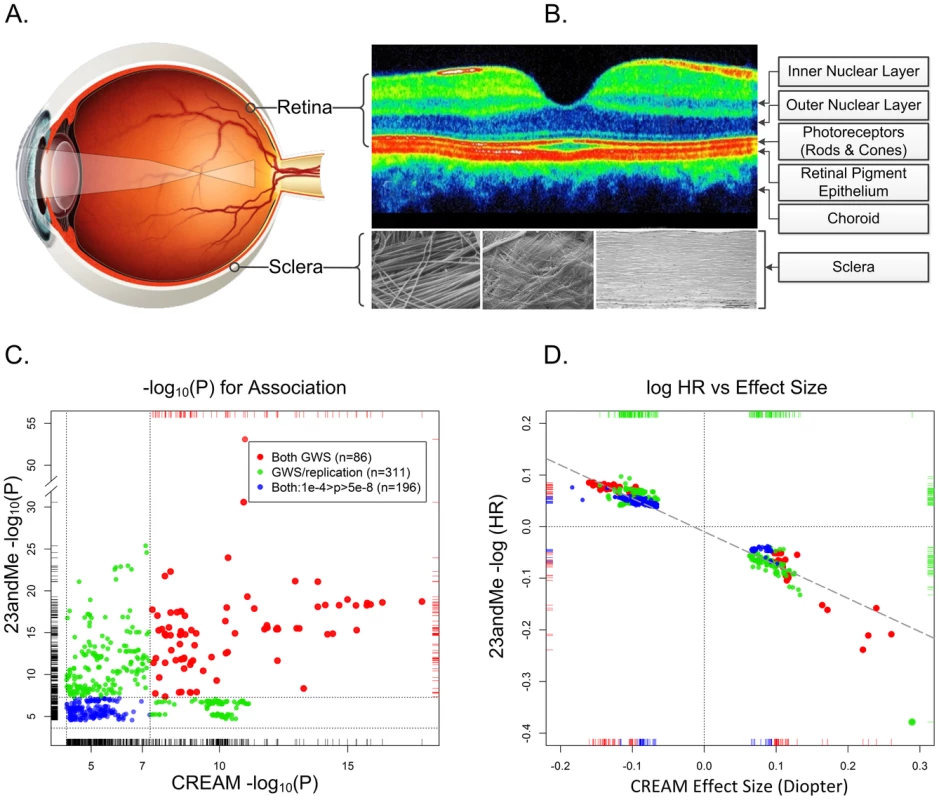
Panel A. The optical basis of myopia. Parallel lights from distantly viewed object focus anterior to the retinal plane, causing blurred distance vision. Although myopia can result from alterations in the optical system (the cornea and lens), myopia is almost always the result of excessive growth of the posterior segment of the eye. This growth occurs through active remodeling of the fibrous sclera. Panel B. Optical coherence tomography cross section of a normal posterior pole. The depression in the middle is the fovea, where the concentration of photoreceptors (and by extension the visual acuity) is greatest. Only four of the ten layers of the neural retina are pointed out. In refractive regulation, visually induced signals are generated by the photoreceptors and processed by other retinal cells (notably, by the amacrine cells, which release neuromodulators and neurotransmitters (including dopamine)). These retinal signals must traverse the retinal pigment epithelium and the highly vascularized choroid in order to regulate eye growth through active remodeling of scleral ECM. The sclera is composed mainly of collagens, but also contains active fibroblasts and normal constituents of ECM (proteoglycans, MMPs, TIMPs, etc.). The composition of the sclera with its organized structure of collagen fibrils are shown under three different magnifications. Panel C. Statistical significance and effect sizes for 23andMe and CREAM studies. Statistical significance of overlapping SNPs with association p-values <1e-04 in both datasets are plotted. Red dots show SNPs that were genomewide significant (GWS; p<5e-08, −log10(p)>7.3) in both studies. Green dots represent SNPs that were genomewide significant in one study and fulfilled our statistical criterion for replication in the other study. Blue dots show SNPs whose p-values for association were between p = 10e-4 and 5e-8 in both datasets. The replication thresholds were the Bonferroni-corrected p-values for a family-wise error rate of 0.05: since the CREAM dataset contained 544 (nonindependent) SNPs that were genomewide significant in 23andMe, the Bonferroni-adjusted p-value threshold for replication was 0.05/544 = 9.2e-05 (or −log10(p) = 4.037). Similarly, there were 308 genomewide significant SNPs in CREAM that were also in the 23andMe dataset presented in Table S2 of Kiefer et al. The replication threshold was therefore 0.05/308 = 2.4e-04 (−log10(p) = 3.615). Dotted horizontal and vertical lines show the thresholds for genomewide significance and replication. Panel D. Natural logarithm of hazard ratios for myopia in 23andMe vs. effect size (beta coefficient) in CREAM. Both analyses assumed an additive effect of the representative allele on the scale used in the analyses. Color coding is identical to the left panel. The dashed diagonal line shows the linear regression of the 23andMe log hazard ratios on CREAM effect sizes. All SNPs are concordant as to the expected direction of their effects (i.e., positive HRs for myopia would be expected to increase the severity of myopia and negative HRs would be expected to decrease the degree of myopia). Dotted lines show the origins where the effects are null. Large GWAS Discovers Numerous Myopia Susceptibility Variants
In a recent issue of PLOS Genetics, Amy K. Kiefer and a group from 23andMe, Inc. [13] reported the results of the largest GWAS (n = 45,771) of a refractive phenotype conducted to date. In addition to confirming previously reported loci [14], [15], they discovered 20 novel loci associated with age of onset of myopia in a European-derived population. Their Cox proportional hazards survival analysis approach proved quite powerful and was ideal for the nature of the data collected: given that the vast majority of myopes develop the condition by early adulthood, ascertainment is effectively complete by age 30. Their findings provide unprecedented insight into the genetics of human myopia and may very well transform our understanding of this perplexing condition. Specifically, the authors identified novel loci containing genes involved in a variety of mechanisms putatively related to visual perception and refractive control, including extracellular matrix (ECM) remodeling, the visual cycle, neuronal development, eye and body growth, retinal cell development, and neural signaling. Their data substantiate the assumption that myopia (more precisely: the myopias) is a complex, multifactorial, and genetically heterogeneous condition. Though their precise roles in refraction regulation are currently unknown, many of these genes' molecular functions and biological processes can be placed within the broader biological context of a retina-to-sclera signaling cascade, similar to the one that maintains optical homeostasis during infancy.
Kiefer et al.'s study was not without some limitations. Most notably, their use of questionnaire data, not validated against medical records or eye exams, likely resulted in substantial misclassification. Moreover, age of onset may not be relevant to the severity of myopia, which is ultimately the main outcome of interest for most clinicians and scientists.
As with most complex diseases, Kiefer et al. found no smoking gun (myopia susceptibility variants did not follow the vision genetics precedent of CFH as the statistical geneticist's equivalent of a slam dunk). Despite highly statistically significant associations, no variant was estimated to have a strong effect on myopia onset: the hazard ratios among the most highly significant SNPs ranged from 0.79 (rs12193446; LAMA2) to 1.15 (rs1381566; LRRC4C). The more common associated polymorphisms had quite modest effects, conferring relative hazard differences of less than 10%. These results suggest that we are pushing the boundaries of the power of traditional GWAS to detect myopia susceptibility genes with reasonably sized studies (although the effect estimates in this study are likely biased toward the null given the misclassification rates expected from questionnaire data). If common variants of large effect did exist, however, Kiefer et al. would surely have found them.
The Three Laws of GWAS: Replication, Replication, Replication
Conventional (i.e., conformist) dogma, in a world of infinitesimal p-values, states that GWAS results are unreliable (minus confirmation through “replication”) no matter the size or statistical power of the “discovery” sample. Kiefer et al. used a validation set of over 8,323 participants who reported on their use of corrective eyewear for myopia before age ten. Using a quite liberal threshold, they statistically replicated ten of their novel loci.
Independent Studies Unequivocally Confirm Their Association Signals
Many readers may be skeptical of replication p-values on the order of 0.01, especially given multiple testing issues. Kiefer et al.'s replication cohort was, to be sure, underpowered to replicate small effects. Nonetheless, their study was, in fact, remarkably successful as revealed by a large independent study: the Consortium for Refractive Error and Myopia (CREAM), of which we are members. CREAM concurrently conducted a meta-analysis of GWAS comprised of 27 studies (n = 37,382) of adults of European descent and five Asian cohorts (n = 8,376) [16]. The study-specific mean ages ranged from 31.4 to 79.9 years, and the average ages in 26 of these studies were >45 years. Unlike the questionnaire data used by Kiefer et al., CREAM phenotypes consisted of direct objective measurements of the quantitative trait, ocular refraction, measured using autorefractometry. Astonishingly, this independent meta-analysis codiscovered genomewide significant association signals (p<5e-08) at 11 of 20 of Kiefer et al.'s “novel” loci (Figure 1, panel C). An additional five loci met our conservative threshold for statistically significant replication based on a Bonferroni correction. In total, 16 of the 20 novel loci identified by Kiefer et al. were confirmed by CREAM; and of the 22 loci discovered by the CREAM analyses, 14 were replicated by 23andMe (Figure 1, panel C). Arguably, nine additional regions could have been added to the list of replicated loci because both studies contained highly significant signals at these loci, albeit at different SNPs.
Perhaps even more surprising was the fact that, although the demographics, analyses, and measurement scales differed markedly between the respective studies, there was a linear relationship between the effect sizes of the statistically significant SNPs common to both studies (Figure 1, panel D). It is quite striking that locus-specific hazard ratios for myopia age of onset (all occurring before age 30) would have predictable, though often small, consequences on the degree of refractive error throughout middle age, and well into the golden years. These relative effects are evident despite substantial changes in refractive error throughout life, in addition to the cumulative influence of a plethora of environmental and behavioral risk factors on refraction.
What's Next?
There is no doubt that the findings published by Kiefer et al. will contribute significantly to our understanding of the processes by which common forms of myopia develop. Their results were strongly (in many cases unequivocally) validated by a large consortium of refractive error GWAS. Survival analysis of population-based samples, however, may not be adequate to detect variants for rare myopia subtypes, such as Mendelian forms of high myopia. For these cases, sequencing of highly ascertained pedigrees should offer better odds of identifying rare causal variants.
Many of the genes identified by the 23andMe and CREAM groups can be linked to biologically plausible pathways for refractive error control. The eye-growth signaling pathway models developed by biologists had previously defined broad constraints on the mechanisms by which ocular development should be controlled by visual inputs. However, scant few genetic animal models for myopia have been developed [17], [18]. Moreover, to our knowledge, none of the refractive error loci identified in the 23andMe and CREAM GWAS have been implicated in visually induced experimental myopia. Hence, the exact mechanisms through which these genes act to regulate ocular refraction are unknown, and current animal models may be inadequate to hypothesize mechanisms for later-onset myopia in humans. The scientific focus will now go back to biology, bioinformatics, and computational biology in an attempt to untangle the effects of these gene networks. Both the 23andMe and CREAM groups have shown that there is no substitute for large sample sizes, and we expect the trend of accumulating ever-larger datasets to continue. Fortunately, refractive error data are easily obtained noninvasively, and Kiefer et al. have shown that age of onset of myopia can be powerful in detecting the signal of refractive error variants.
Future studies should not, however, neglect the large effect of extrinsic factors on refractive error. A significant body of work has linked refractive errors with a number of environmental, ecological, and behavioral risk factors (summarized in, e.g., [6], [19]). In fact, societies in which these putative risk factors are uncommon have low prevalences of myopia [20]–[23]. This suggests that, despite high heritability, exposure to deleterious environmental influences (i.e., reading and near work) may be essential for refractive variation to be manifested. Hence, a clearer picture of myopia and refractive error genetics will require incorporating measures of environmental exposures into our statistical analyses in GWAS. Finally, it is imperative to conduct whole-genome searches in a variety of populations and ethnicities, not only to further validate these results, but to investigate the causes of the wide disparities in the distribution of ocular refraction within, and among, these groups.
Zdroje
1. VitaleS, SperdutoRD, FerrisFL3rd (2009) Increased prevalence of myopia in the United States between 1971–1972 and 1999–2004. Arch Ophthalmol 127 : 1632–1639.
2. LinLL, ShihYF, HsiaoCK, ChenCJ (2004) Prevalence of myopia in Taiwanese schoolchildren: 1983 to 2000. Ann Acad Med Singapore 33 : 27–33.
3. KempenJH, MitchellP, LeeKE, TielschJM, BromanAT, et al. (2004) The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol 122 : 495–505.
4. WuHM, SeetB, YapEP, SawSM, LimTH, et al. (2001) Does education explain ethnic differences in myopia prevalence? A population-based study of young adult males in Singapore. Optom Vis Sci 78 : 234–239.
5. HeM, ZengJ, LiuY, XuJ, PokharelGP, et al. (2004) Refractive error and visual impairment in urban children in southern China. Invest Ophthalmol Vis Sci 45 : 793–799.
6. WojciechowskiR (2011) Nature and nurture: the complex genetics of myopia and refractive error. Clin Genet 79 : 301–320.
7. Kepler J (1604) Ad Vitellionem paralipomena, quibus astronomiae pars optica traditur… Tractatum luculentum de modo visionis, & humorum oculi usu, contra opticos & anatomicos. Frankfurt: Claudius Marnius & heirs of Joannes Aubrius.
8. WorthC (1906) Hereditary influence in myopia. Trans Opthal Soc U K 26 : 141–144.
9. WoldKC (1949) Hereditary myopia. Arch Ophthalmol 42 : 225–237.
10. ShermanSM, NortonTT, CasagrandeVA (1977) Myopia in the lid-sutured tree shrew (Tupaia glis). Brain Res 124 : 154–157.
11. WallmanJ, TurkelJ, TrachtmanJ (1978) Extreme myopia produced by modest change in early visual experience. Science 201 : 1249–1251.
12. WieselTN, RaviolaE (1977) Myopia and eye enlargement after neonatal lid fusion in monkeys. Nature 266 : 66–68.
13. KieferAK, TungJY, DoCB, HindsDA, MountainJL, et al. (2013) Genome-wide analysis points to roles for extracellular matrix remodeling, the visual cycle, and neuronal development in myopia. PLoS Genet 9 (2) e1003299 doi:10.1371/journal.pgen.1003299.
14. HysiPG, YoungTL, MackeyDA, AndrewT, Fernandez-MedardeA, et al. (2010) A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet 42 : 902–905.
15. SoloukiAM, VerhoevenVJ, van DuijnCM, VerkerkAJ, IkramMK, et al. (2010) A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet 42 : 897–901.
16. VerhoevenV, HysiP, WojciechowskiR, FanQ, GuggenheimJ, et al. (2013) Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat Genet 45 : 314–318.
17. ChenYP, HockingPM, WangL, PovazayB, PrasharA, et al. (2011) Selective breeding for susceptibility to myopia reveals a gene-environment interaction. Invest Ophthalmol Vis Sci 52 : 4003–4011.
18. TroiloD, LiT, GlasserA, HowlandHC (1995) Differences in eye growth and the response to visual deprivation in different strains of chicken. Vision Res 35 : 1211–1216.
19. MorganI, RoseK (2005) How genetic is school myopia? Prog Retin Eye Res 24 : 1–38.
20. PokharelGP, NegrelAD, MunozSR, EllweinLB (2000) Refractive error study in children: results from Mechi Zone, Nepal. Am J Ophthalmol 129 : 436–444.
21. CassonRJ, KahawitaS, KongA, MueckeJ, SisaleumsakS, et al. (2012) Exceptionally low prevalence of refractive error and visual impairment in schoolchildren from Lao People's Democratic Republic. Ophthalmology 119 : 2021–2027.
22. PeetJA, CotchMF, WojciechowskiR, Bailey-WilsonJE, StambolianD (2007) Heritability and familial aggregation of refractive error in the Old Order Amish. Invest Ophthalmol Vis Sci 48 : 4002–4006.
23. YoungFA, LearyGA, BaldwinWR, WestDC, BoxRA, et al. (1969) The transmission of refractive errors within eskimo families. Am J Optom Arch Am Acad Optom 46 : 676–685.
Štítky
Genetika Reprodukční medicína
Článek The G4 GenomeČlánek Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance inČlánek RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations inČlánek Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during DevelopmentČlánek Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic LethalityČlánek DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 4
-
Všechny články tohoto čísla
- Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
- A Big Catch for Germ Cell Tumour Research
- The Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
- A Nonsynonymous Polymorphism in as a Risk Factor for Human Unexplained Cardiac Arrest with Documented Ventricular Fibrillation
- The Hourglass and the Early Conservation Models—Co-Existing Patterns of Developmental Constraints in Vertebrates
- Smaug/SAMD4A Restores Translational Activity of CUGBP1 and Suppresses CUG-Induced Myopathy
- Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
- The G4 Genome
- Extensive Natural Epigenetic Variation at a Originated Gene
- Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases
- The Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
- TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
- Aconitase Causes Iron Toxicity in Mutants
- Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
- Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
- Sex-Differential Selection and the Evolution of X Inactivation Strategies
- Identification of a Tissue-Selective Heat Shock Response Regulatory Network
- Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity
- RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in
- Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
- Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
- ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair
- Iron-Responsive miR-485-3p Regulates Cellular Iron Homeostasis by Targeting Ferroportin
- Mutations in Predispose Zebrafish and Humans to Seminomas
- Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands
- Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during Development
- All SNPs Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated SNPs
- Functional 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- Genetic Requirements for Signaling from an Autoactive Plant NB-LRR Intracellular Innate Immune Receptor
- SNF5 Is an Essential Executor of Epigenetic Regulation during Differentiation
- Dialects of the DNA Uptake Sequence in
- Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate–Invertebrate Gap
- Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing
- High-Resolution Mapping of Spontaneous Mitotic Recombination Hotspots on the 1.1 Mb Arm of Yeast Chromosome IV
- Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
- Evolution after Introduction of a Novel Metabolic Pathway Consistently Leads to Restoration of Wild-Type Physiology
- Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
- Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
- Signatures of Diversifying Selection in European Pig Breeds
- The Chromosomal Passenger Protein Birc5b Organizes Microfilaments and Germ Plasm in the Zebrafish Embryo
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- Regulates Synaptic Development and Endocytosis by Suppressing Filamentous Actin Assembly
- Sensory Neuron-Derived Eph Regulates Glomerular Arbors and Modulatory Function of a Central Serotonergic Neuron
- Analysis of Rare, Exonic Variation amongst Subjects with Autism Spectrum Disorders and Population Controls
- Scavenger Receptors Mediate the Role of SUMO and Ftz-f1 in Steroidogenesis
- DNA Double-Strand Breaks Coupled with PARP1 and HNRNPA2B1 Binding Sites Flank Coordinately Expressed Domains in Human Chromosomes
- High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells
- Comparative Genomics of and the Bacterial Species Concept
- Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming
- Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
- The Genetic Correlation between Height and IQ: Shared Genes or Assortative Mating?
- Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
- An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
- The Genome Organization of Reflects Its Lifestyle
- Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood
- Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate
- Site-Specific Phosphorylation of the DNA Damage Response Mediator Rad9 by Cyclin-Dependent Kinases Regulates Activation of Checkpoint Kinase 1
- Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin
- Identification of , a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis
- Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
- DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
- Asynchronous Replication, Mono-Allelic Expression, and Long Range -Effects of
- Differential Association of the Conserved SUMO Ligase Zip3 with Meiotic Double-Strand Break Sites Reveals Regional Variations in the Outcome of Meiotic Recombination
- Focusing In on the Complex Genetics of Myopia
- Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans
- Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types
- Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD
- Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse
- Genetic and Genomic Architecture of the Evolution of Resistance to Antifungal Drug Combinations
- Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs
- Functional Dissection of the Condensin Subunit Cap-G Reveals Its Exclusive Association with Condensin I
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The G4 Genome
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



