-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaGenome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
Some sheep breeds are naturally prolific, and they are very informative for the studies of reproductive genetics and physiology. Major genes increasing litter size (LS) and ovulation rate (OR) were suspected in the French Grivette and the Polish Olkuska sheep populations, respectively. To identify genetic variants responsible for the highly prolific phenotype in these two breeds, genome-wide association studies (GWAS) followed by complementary genetic and functional analyses were performed. Highly prolific ewes (cases) and normal prolific ewes (controls) from each breed were genotyped using the Illumina OvineSNP50 Genotyping Beadchip. In both populations, an X chromosome region, close to the BMP15 gene, harbored clusters of markers with suggestive evidence of association at significance levels between 1E−05 and 1E−07. The BMP15 candidate gene was then sequenced, and two novel non-conservative mutations called FecXGr and FecXO were identified in the Grivette and Olkuska breeds, respectively. The two mutations were associated with the highly prolific phenotype (pFecXGr = 5.98E−06 and pFecXO = 2.55E−08). Homozygous ewes for the mutated allele showed a significantly increased prolificacy (FecXGr/FecXGr, LS = 2.50±0.65 versus FecX+/FecXGr, LS = 1.93±0.42, p<1E−03 and FecXO/FecXO, OR = 3.28±0.85 versus FecX+/FecXO, OR = 2.02±0.47, p<1E−03). Both mutations are located in very well conserved motifs of the protein and altered the BMP15 signaling activity in vitro using a BMP-responsive luciferase test in COV434 granulosa cells. Thus, we have identified two novel mutations in the BMP15 gene associated with increased LS and OR. Notably, homozygous FecXGr/FecXGr Grivette and homozygous FecXO/FecXO Olkuska ewes are hyperprolific in striking contrast with the sterility exhibited by all other known homozygous BMP15 mutations. Our results bring new insights into the key role played by the BMP15 protein in ovarian function and could contribute to a better understanding of the pathogenesis of women′s fertility disorders.
Published in the journal: . PLoS Genet 9(4): e32767. doi:10.1371/journal.pgen.1003482
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003482Summary
Some sheep breeds are naturally prolific, and they are very informative for the studies of reproductive genetics and physiology. Major genes increasing litter size (LS) and ovulation rate (OR) were suspected in the French Grivette and the Polish Olkuska sheep populations, respectively. To identify genetic variants responsible for the highly prolific phenotype in these two breeds, genome-wide association studies (GWAS) followed by complementary genetic and functional analyses were performed. Highly prolific ewes (cases) and normal prolific ewes (controls) from each breed were genotyped using the Illumina OvineSNP50 Genotyping Beadchip. In both populations, an X chromosome region, close to the BMP15 gene, harbored clusters of markers with suggestive evidence of association at significance levels between 1E−05 and 1E−07. The BMP15 candidate gene was then sequenced, and two novel non-conservative mutations called FecXGr and FecXO were identified in the Grivette and Olkuska breeds, respectively. The two mutations were associated with the highly prolific phenotype (pFecXGr = 5.98E−06 and pFecXO = 2.55E−08). Homozygous ewes for the mutated allele showed a significantly increased prolificacy (FecXGr/FecXGr, LS = 2.50±0.65 versus FecX+/FecXGr, LS = 1.93±0.42, p<1E−03 and FecXO/FecXO, OR = 3.28±0.85 versus FecX+/FecXO, OR = 2.02±0.47, p<1E−03). Both mutations are located in very well conserved motifs of the protein and altered the BMP15 signaling activity in vitro using a BMP-responsive luciferase test in COV434 granulosa cells. Thus, we have identified two novel mutations in the BMP15 gene associated with increased LS and OR. Notably, homozygous FecXGr/FecXGr Grivette and homozygous FecXO/FecXO Olkuska ewes are hyperprolific in striking contrast with the sterility exhibited by all other known homozygous BMP15 mutations. Our results bring new insights into the key role played by the BMP15 protein in ovarian function and could contribute to a better understanding of the pathogenesis of women′s fertility disorders.
Introduction
There is strong evidence supporting the concept that oocyte plays a central role in follicle growth and developmental regulation [1], [2], [3]. It has been established that ovary-derived transforming growth factor-ß (TGFß) family members play an integral role during folliculogenesis. Indeed, the two paracrine factors Bone Morphogenetic Protein 15 (BMP15) and Growth and Differentiation Factor 9 (GDF9) stimulate follicle growth [4], [5], promote granulosa cell proliferation [6], [7], influence cell-survival signaling [8], [9] and modulate other growth factors and hormones [10], [11], [12].
The identification of BMP15, GDF9 and Bone Morphogenetic Protein Receptor 1B (BMPR1B) gene mutations (Table 1) as the causal mechanism underlying either the highly prolific or infertile phenotypes of several sheep breeds in a dosage-sensitive manner highlighted the crucial role these genes play in ovarian function. The TGFß family member BMP15 was the first gene to be associated with prolificacy. All the known BMP15 mutations (named FecXI,H,B,G,L or R) produce the same phenotype i.e. heterozygous ewes are highly prolific whereas homozygous females are infertile due to a blockade of follicular development at the primary stage [13], [14], [15], [16], [17]. A second member of the BMP pathway, BMPR1B, was later found to be associated with prolificacy. The genetic variant (FecBB) segregating in the Booroola Merinos breed presents an additive effect on ovulation rate (OR) and a partially dominant effect on litter size (LS) [18], [19], [20]. For GDF9 (known as FecG), also member of the TGFß family, the two first described mutations present a phenotypic inheritance pattern similar to all BMP15 variants [14], [21]. A third mutation in GDF9 (FecGE) associated with prolificacy was identified recently in the Brazilian Santa Inês strain [22]. Interestingly, Silva et al. showed for the first time a novel phenotype since FecGE homozygous ewes are not sterile but exhibit a significant increase, compared to non mutated individuals, in their OR (2.22±0.12 vs. 1.22±0.11) and LS (1.78 vs. 1.13) [22].
Tab. 1. Genetics variants causing prolificacy phenotypes in sheep. 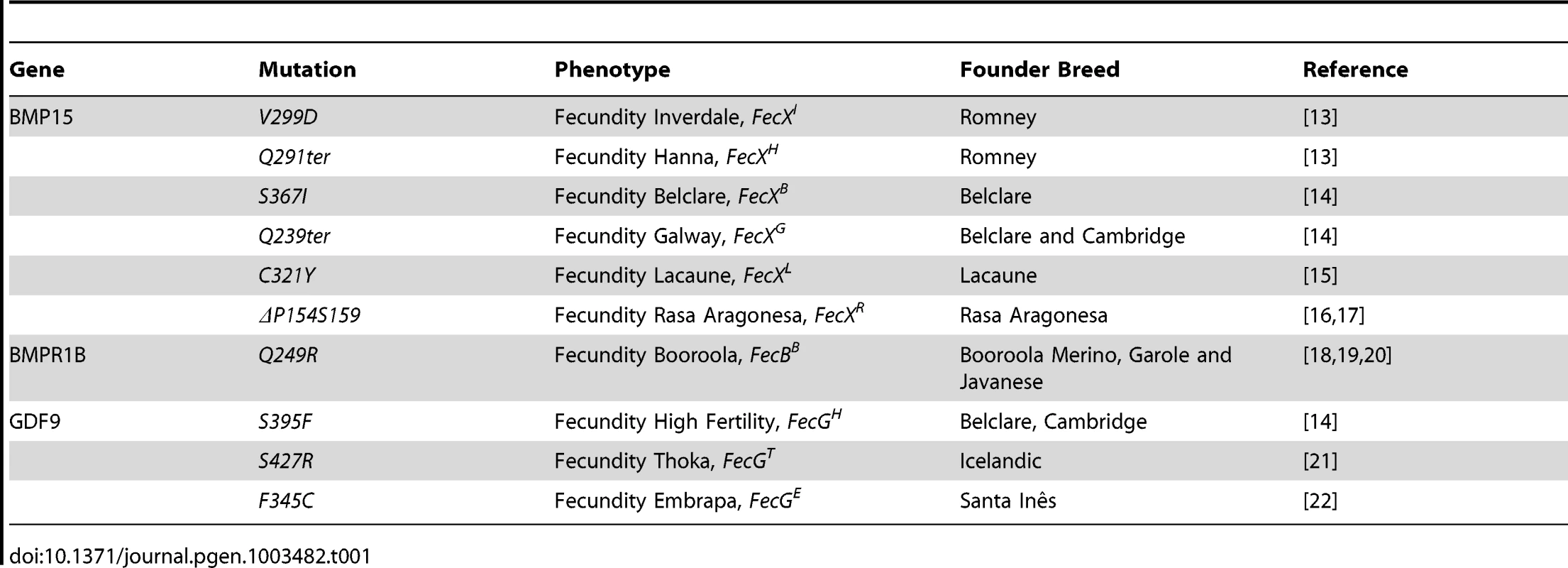
Similarly to sheep, a large number of mutations in the GDF9 and BMP15 genes have been described in women with fertility disorders. The identification of GDF9 missense and nonsense mutations in premature ovarian failure (POF) patients [23], [24], [25] but also in mothers of dizygotic twins [26], [27], [28] suggests that altered GDF9 function is involved in ovarian dysfunction and polyovulatory phenotypes. Indeed, 3 missense variants GDF9P103S, GDF9P374L and GDF9R454C were significantly associated with an increased OR in women [27]. Critical roles of BMP15 in female fertility have also been demonstrated in women. The first BMP15 mutation (BMP15Y235C) responsible for hypergonadotropic ovarian failure, due to ovarian dysgenesis, was detected in two sisters who inherited the variant from their father [29]. A number of other mutations and rare deletions in those 2 genes have been described in women with POF [24], [29], [30], [31], [32]. Recently, Hanevik et al. [33] confirmed results previously observed by Moron et al. [34] showing that a BMP15 variant (BMP15−9G) was associated with ovarian hyperstimulation syndrome (OHSS), similar to effects observed in ewes with heterozygous BMP15 mutations. Therefore, GDF9 and BMP15 are likely to alter OR and LS in women as well as in ewes. Interestingly, BMP15 regulates ovulation rate and female fertility in a species-specific manner, being crucial in humans and sheep and largely trivial in mice since loss-of-function of BMP15 results only in subfertility [35].
Although numerous mutations improving reproduction traits have been discovered in various sheep strains, the genetic variants have still to be identified for some other prolific sheep breeds. The French Grivette and the Polish Olkuska breeds present good maternal characteristics and abnormally high LS and OR, respectively. Moreover, both populations exhibit a very high variability of mean prolificacy among related females, suggesting the existence of an autosomal major gene for prolificacy segregating the same way as the FecBB mutation (reviewed in [36]). The FecBB or FecXI mutations do not account for the OR phenotype in Olkuska ewes [37]. This work aims at identifying genetic variants affecting LS and OR phenotypes in the French Grivette and Polish Olkuska sheep populations, respectively. A GWAS based on a case/control design followed by fine-mapping genetic analyses was performed. We identified 2 novel non-conservative mutations in the BMP15 gene associated with an increase of LS and OR. Following the nomenclature used for previous fecundity genes, these 2 mutations were named FecXGr in the Grivette population and FecXO in the Olkuska population. Noteworthy, homozygous ewes of both strains are highly prolific but not sterile as known so far. Our results bring new insights into the key role played by the BMP15 protein in ovarian function.
Results
Genetic association analyses
To identify loci associated with highly prolific phenotype in the French Grivette and the Polish Olkuska sheep breeds, a GWAS comparing allele frequencies between highly prolific ewes (cases) versus normally prolific ewes (controls) was conducted as outlined in Materials and Methods and Table S1. Genotype data from the Illumina OvineSNP50 Genotyping Beadchip was obtained for 39 ewes (28 cases vs. 11 controls) and 63 ewes (29 cases vs. 34 controls) from the Grivette and the Olkuska populations, respectively. No significant genome-wide association after Bonferroni correction was found in the French Grivette breed although an association was suggested for a cluster of markers located on the X chromosome (Figure 1A). Indeed, 5 OARX SNPs, in a region close to BMP15 gene, present evidence of association at chromosome-wide level as shown on Figure 1C and in Table 2. For the Polish Olkuska breed, 2 and 4 markers located on OARX close to the BMP15 gene are significantly associated with OR at genome-wide and chromosome-wide levels, respectively (Figure 1B, 1D and Table 2).
Fig. 1. Genome-wide and chromosome-wide association results. 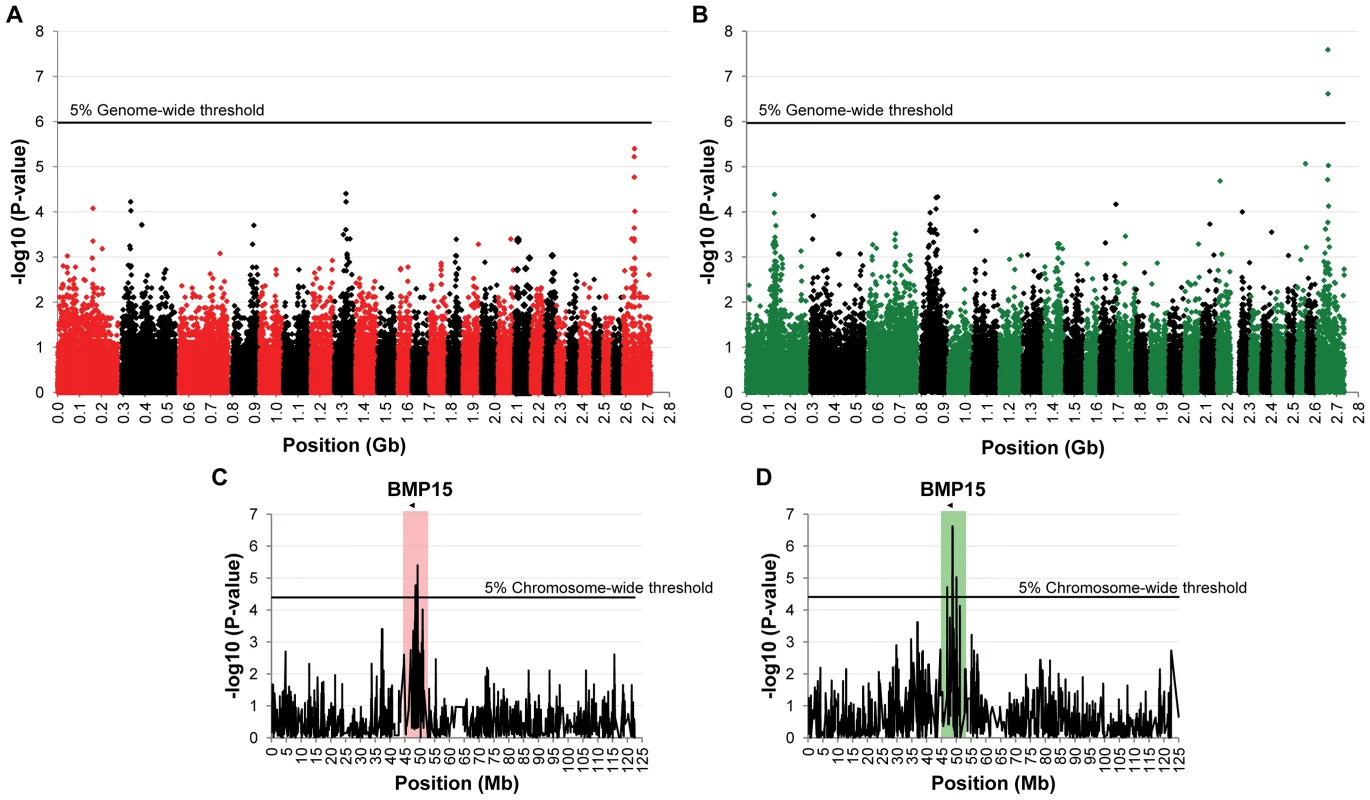
(A) Genome-wide association results for litter size in the French Grivette sheep population. (B) Genome-wide association results for ovulation rate in the Polish Olkuska sheep population. Manhattan plots show the combined association signals (−log10(p)) on the y-axis versus SNPs position in the sheep genome on the x-axis and ordered by chromosome number (OARv2.0 available on http://www.livestockgenomics.csiro.au/sheep/ website). Black lines represent the 5% genome-wide threshold. Chromosomes are ordered from OAR1 to OAR26 and the X chromosome is the last one. (C) OARX chromosome-wide association results for litter size in the French Grivette sheep population. (D) OARX chromosome-wide association results for ovulation rate in the Polish Olkuska sheep population. Manhattan plots show the combined association signals (−log10(p)) on the y-axis versus SNPs position on the X chromosome (OARv2.0 available on http://www.livestockgenomics.csiro.au/sheep/ website). Black lines represent the 5% chromosome-wide threshold. Red and green boxes pinpoint locus where significant association results are obtained for several markers in the Grivette and Olkuska populations, respectively. The location of the BMP15 gene is mentioned by an arrow. Tab. 2. Markers significantly associated with prolificacy. 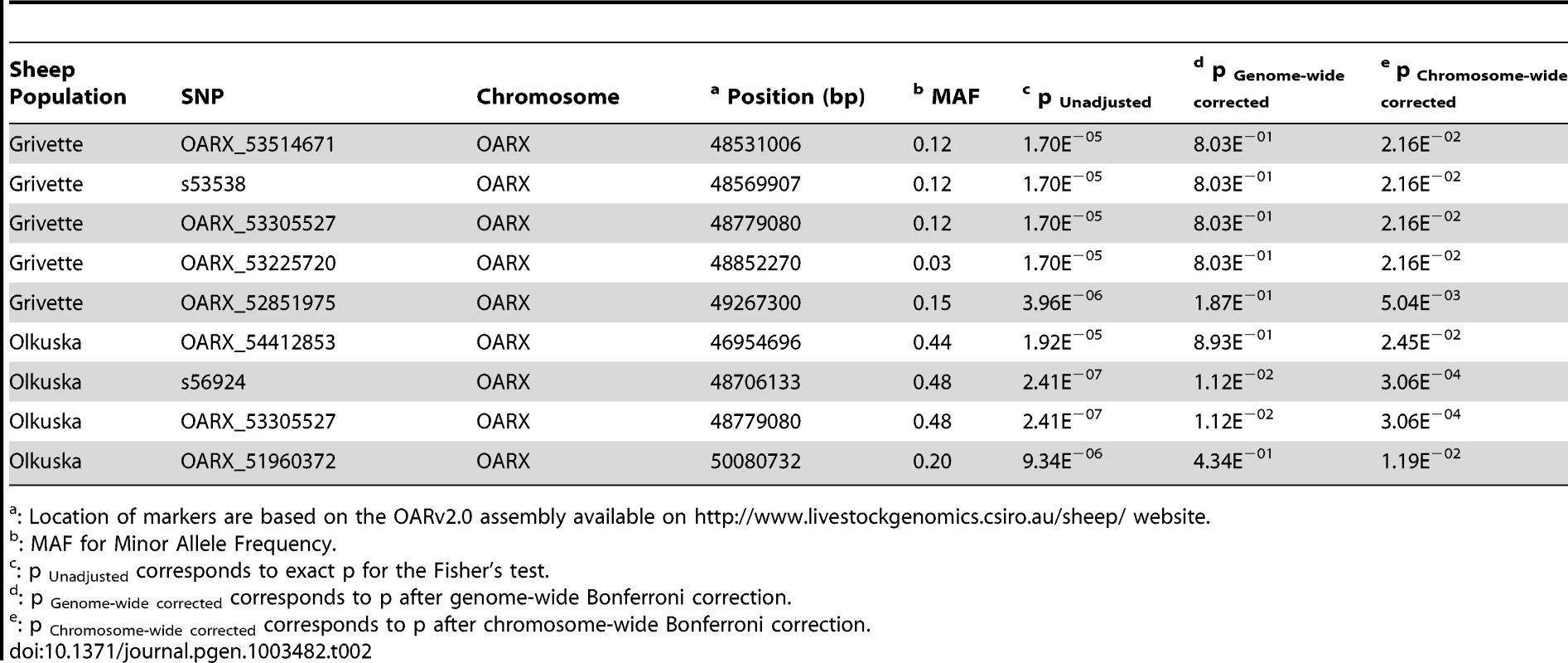
: Location of markers are based on the OARv2.0 assembly available on http://www.livestockgenomics.csiro.au/sheep/ website. To further characterize the X chromosome locus, we selected 87 markers spanning the 10 Mb region (from 45 Mb to 55 Mb according to the OARv2.0 assembly available on (http://www.livestockgenomics.csiro.au/sheep/ website) and we determined the most likely linkage phase for each individual. After haplotype clusterization for each population, we identified a specific segment that was more frequent in highly prolific ewes than in control individuals as shown on Figure 2A and 2B. In the French Grivette population, a significant association was found for the 1.3 Mb (48240883 bp–49526964 bp) haplotype containing 20 markers (Frcases = 0.95 vs. Frcontrols = 0.54, pcorrected = 9.90E−03) (Figure 2A). Similarly, a different haplotype but located in the same region (48240883 bp–49381143 bp) was shown significantly associated with the OR phenotype in the Polish Olkuska breed (Frcases = 0.76 vs. Frcontrols = 0.27, corrected = 9.90E−04) (Figure 2B).
Fig. 2. Clusterization of haplotypes reconstructed at the OARX locus. 
(A) Haplotypes determined in the French Grivette sheep population. (B) Haplotypes determined in the Polish Olkuska sheep population. 87 markers located in the interest OARX region (45 Mb–55 Mb) were selected to reconstruct haplotypes. Each column represents one SNP and each line represents one haplotype. For one marker (i) allele 1 is in red (Grivette) or green (Olkuska) in controls, respectively or black in cases, (ii) allele 2 is in white when the phase was unambiguous and (iii) dark grey colour represents unphased SNP. Haplotypes were ordered to distinguish controls versus cases and clusterized to classify similar clades of haplotypes. Markers with evidence of association at significance levels are marked with a star or a hash in French Grivette and Polish Olkuska sheep populations, respectively. In both breeds, the specific haplotype preferentially selected in highly prolific ewes (cases) is symbolized by red (Grivette) and green (Olkuska) boxes. The BMP15 gene (48140251 bp–48146740 bp) is located between markers named OARX_6082722 and OARX_56342973. Characterization of mutations
The very close location of the identified region to the BMP15 gene (48140251 bp–48146740 bp) and the crucial role of BMP15 in ovarian function prompted us to consider BMP15 as a positional and functional candidate gene. The BMP15 coding sequence was obtained for all the individuals genotyped in GWAS for both sheep breeds.
In the French Grivette population, 3 polymorphisms were identified: the already known c.28–30delCTT and c.747C>T and a new c.950C>T leading to the ΔL10 deletion, the P249P synonymous substitution and a non-conservative (T317I) substitution (Figure S1A), respectively as shown in Table 3. The distribution of the BMP15T317I mutation strongly suggested its association with increased litter size since 26 out of the 28 highly prolific ewes were homozygous for the T mutated allele (Table 3). To validate this hypothesis, the BMP15T317I genotype was added to the GWAS and a significant chromosome-wide association was found (punadjusted = 5.98E−06, pGenome-wide corrected = 2.83E−01 and pChromosome-wide corrected = 7.61E−03) (Figure S2A). Preliminary genetic results were confirmed via the genotyping of 360 additional ewes randomly chosen in 9 flocks and then litter size means between the 3 genotypes at BMP15T317I (C/C vs. C/T vs. T/T) were compared as shown on Figure 3A. Litter size of individuals with the T/T genotype (n = 119, LS = 2.50±0.65) was significantly higher than litter sizes of ewes with the C/C (n = 85, LS = 1.83±0.41) or the C/T (n = 195, LS = 1.93±0.42) genotypes (p<1E−03). However, there was no difference between LS from C/T and C/C genotypes although a trend was observed (p = 0.06). These results were confirmed on the ovulation rate measured for a few ewes (n = 27) (Figure 3C). Additionally, 6 kb of the 5′ regulatory region of the BMP15 gene were sequenced in 2 extreme ewes (homozygous C/C, LS = 1.17 and homozygous T/T, LS = 4.33) and no polymorphism was identified, therefore reinforcing the role of the BMP15T317I substitution.
Fig. 3. Genotypic distributions of FecXGr and FecXO mutations. 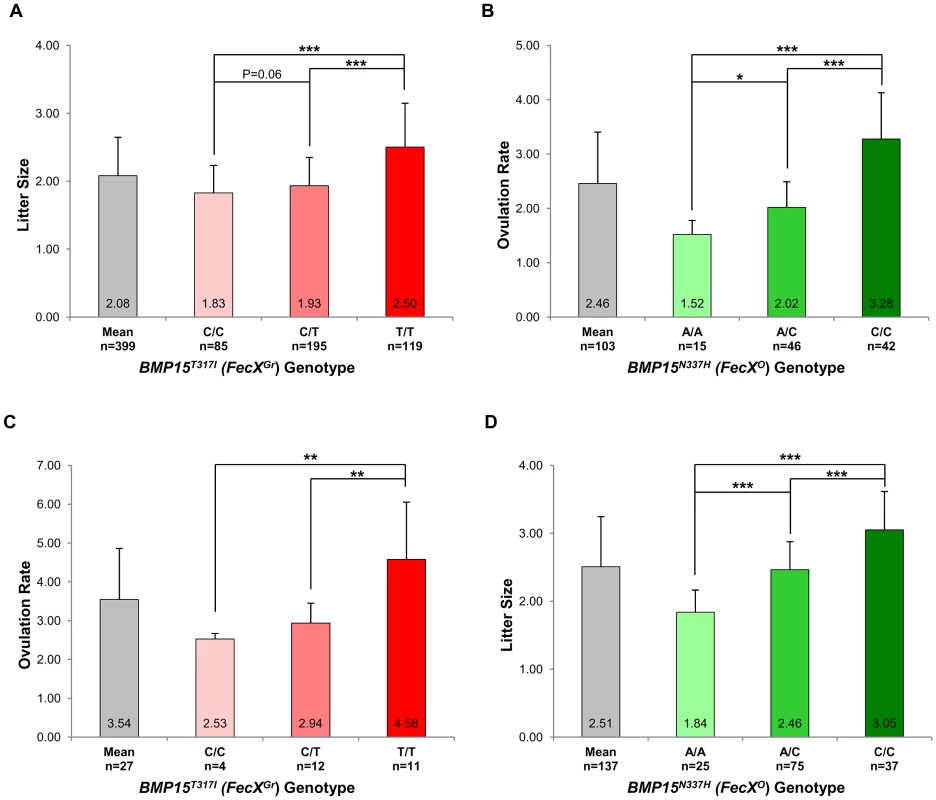
(A) Genotypic distribution of the BMP15T317I (FecXGr) in the French Grivette sheep population for the litter size phenotype. (B) Genotypic distribution of the BMP15N337H (FecXO) in the Polish Olkuska sheep population for the ovulation rate phenotype. (C) Genotypic distribution of the BMP15T317I (FecXGr) in the French Grivette sheep population for the ovulation rate phenotype. (D) Genotypic distribution of the BMP15N337H (FecXO) in the Polish Olkuska sheep population for the litter size phenotype. The means LS or OR in breeds are firstly presented then ewes were ordered according to their genotype at the mutation of interest. Means±SD for prolificacy were calculated for the 3 groups of genotype and are noted into each histogram bar. Number of ewes counted per group of genotype is mentioned (n). Pairwise statistical comparisons using a one-way ANOVA test between means of genotype's clades were performed and results of statistic test are symbolized by stars. p: * = p<5E−02; ** = p<1E−02 and *** = p<1E−03. Tab. 3. BMP15 genetic variants identified in the Grivette and the Olkuska populations. 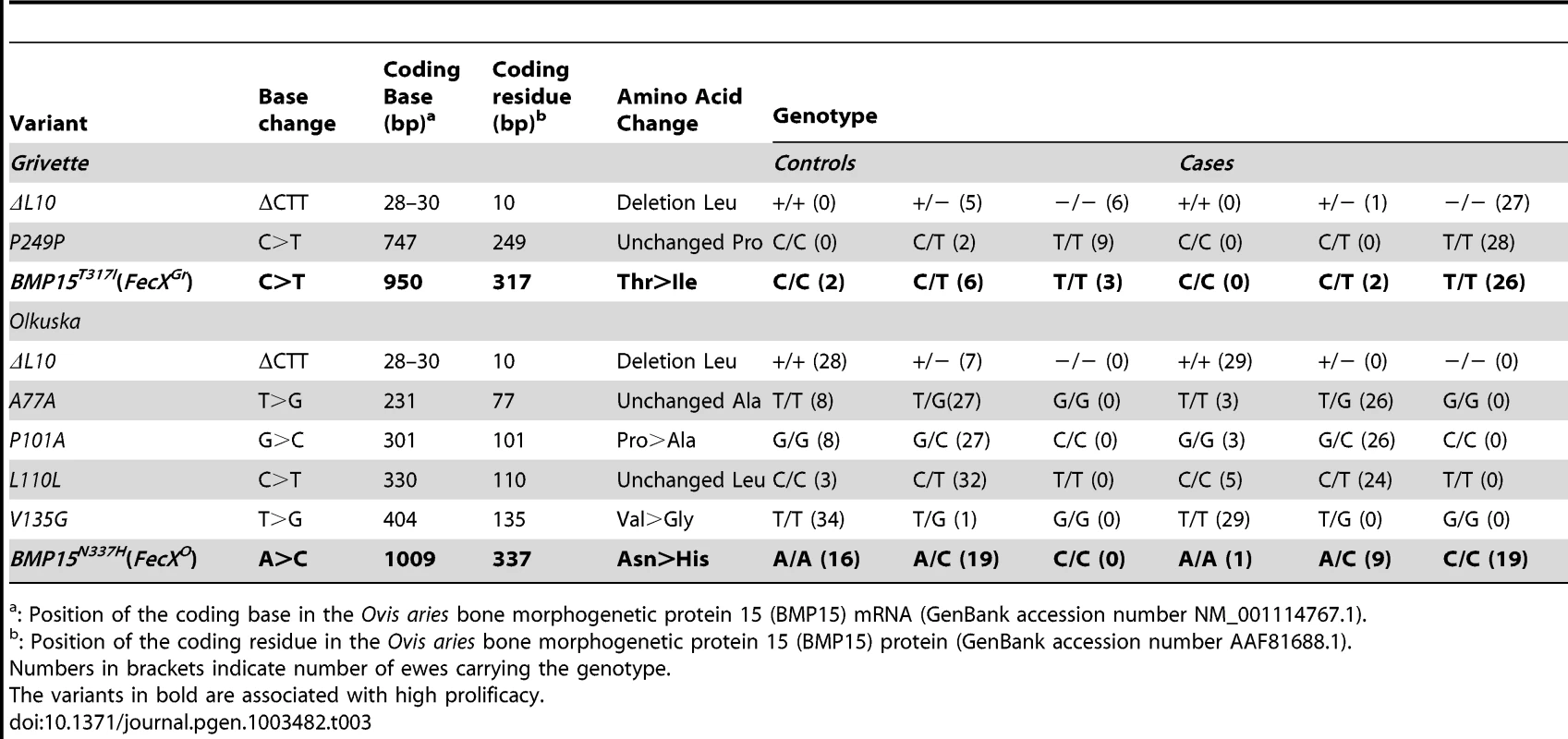
: Position of the coding base in the Ovis aries bone morphogenetic protein 15 (BMP15) mRNA (GenBank accession number NM_001114767.1). A similar strategy was used for the Polish Olkuska breed, and 6 polymorphisms were identified including 5 new ones. Among them, c.231C>T and c.330C>T led to synonymous substitutions (A77A and L110L), and c.301G>C, c.404T>G and c.1009A>C cause P101A, V135G and N337H non-conservative substitutions (Figure S1B), respectively (Table 3). Only the BMP15N337H mutation was shown to be more frequent in cases than in controls (0.81 vs. 0.26) (Table 3) and was significantly associated to OR at a genome-wide level (punadjusted = 2.55E−08, pGenome-wide corrected = 1.18E−03) (Figure S2B). Analysis of the genotypic distribution of the BMP15N337H polymorphism in additional Olkuska ewes confirmed the key role of the mutation on OR. Indeed, the 3 groups of genotype showed statistically different ovulation rate means (A/A, OR = 1.52±0.26 vs. A/C, OR = 2.02±0.47, p<5E−02; A/A, OR = 1.52±0.26 vs. C/C, OR = 3.28±0.85, p<1E−03 and A/C, OR = 2.02±0.47 vs. C/C, OR = 3.28±0.85, p<1E−03) as shown on Figure 3B. These results were validated on the litter size phenotype (Figure 3D). Polymorphisms were identified in the 6 kb 5′ regulatory region of the BMP15 gene of 4 extreme animals but none of them seemed to be associated with the high prolificacy emphasizing the interest of the BMP15N337H substitution (Table S2).
Altogether, this data suggests that the non-conservative BMP15T317I and BMP15N337H mutations, called FecXGr and FecXO, respectively, are causal mutations. Both mutations are associated with increased litter size and ovulation rate in the French Grivette and Polish Olkuska sheep populations. Interestingly, homozygous ewes are highly prolific and not sterile, a phenotype discordant from all BMP15 mutations described so far in sheep.
Functional effects of mutations
Both FecXGr and FecXO mutations are closely located into two very well conserved domains of the sheep, cow, pig, human and mouse BMP15 proteins (Figure 4). Impacts of BMP15T317I and BMP15N337H variants on the intrinsic properties of the BMP15 protein were estimated by comparing hydrophobicity, polarity and molecular weight between mutated and wild type BMP15 proteins. FecXGr, which corresponds to a substitution of a threonine to an isoleucine, clearly affected the hydrophobicity of the protein while FecXO altered the polarity and the molecular weight of the protein by replacing an asparagine to a histidine as shown on Figure S3.
Fig. 4. BMP15 multi-species sequences alignment and position of sheep mutations. 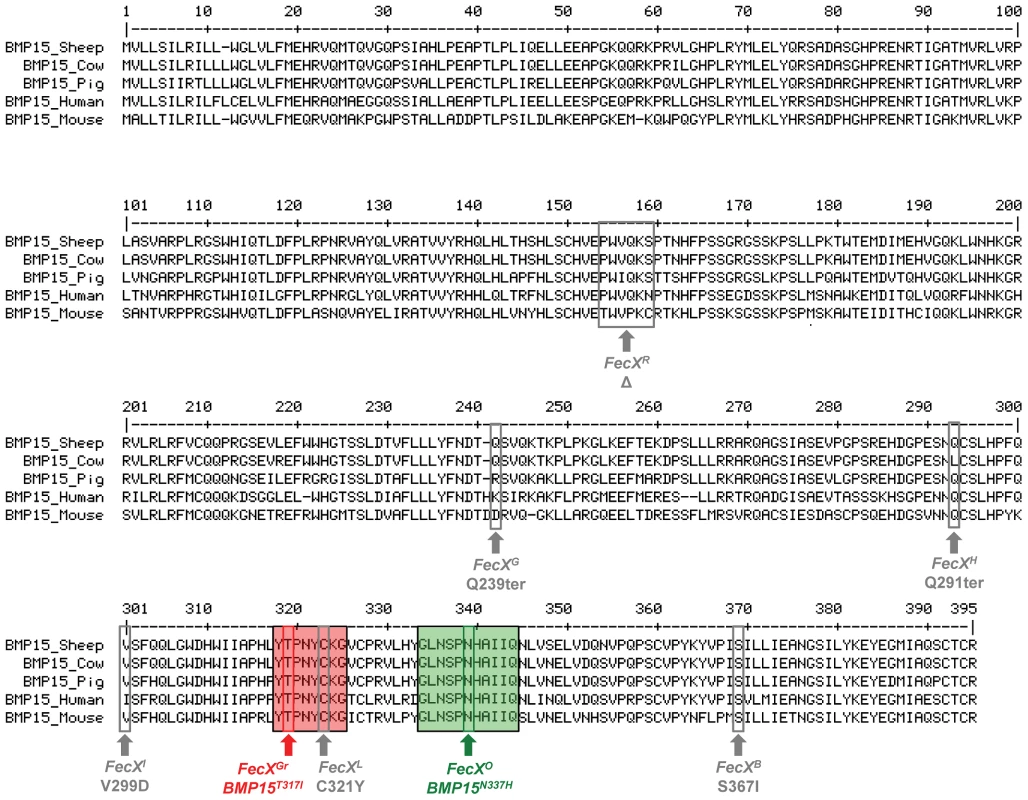
The BMP15 protein sequences from Ovis aries (GenBank AAF81688.1), Bos taurus (GenBank DAA12832.1), Sus scrofa (GenBank NP_001005155.1), Mus musculus (GenBank NP_033887.1) and Homo sapiens (GeneBank NP_005439.2) were aligned and compared. Grey boxes and arrows represent known mutations as reviewed in Otsuka et al. [12]. FecXGr and FecXO mutations are in red or green frames, respectively, themselves into bigger frames symbolizing conserved protein motifs. To evaluate the effects and then the causality of these two naturally occurring mutations on the BMP15 signaling activity, each mutated allele was introduced by targeted mutagenesis in the human BMP15 expressing plasmid. The functional assay was realized in vitro using a human granulosa-derived COV434 cell line stably expressing a BMP-responsive element luciferase reporter construct, as previously described [32]. The quantification of the luciferase activity after transient transfection was compared between plasmids expressing the wild-type allele, the mutated allele and a mix of both, mimicking the heterozygous status (Figure 5). As positive controls, COV434 cells expressing the luciferase reporter gene showed a significant activation of the BMP signaling pathway when stimulated with 100 ng of the exogenous recombinant human BMP15 (p<1E−03), or when transiently transfected with the wild-type human BMP15 expressing construct (p<1E−03). In comparison to the wild-type, the BMP15T317I mutation totally abolished the BMP15 signaling activity. When the BMP15T317I plasmid was cotransfected with an equal amount of the wild-type form, the impaired luciferase activity was partially rescued. Concerning the BMP15N337H mutation, it altered also the BMP15 signaling by half-reducing its activity (p<1E−02) and this was totally rescued by the BMP15T317I/WT cotransfection. This data shows that the two novel mutations drastically affect the signaling pathway of BMP15 and thus, supports their causal role into the highly prolific phenotype observed in the French Grivette and Polish Olkuska sheep populations.
Fig. 5. Functional effects of FecXGr and FecXO mutations on the BMP15 activity. 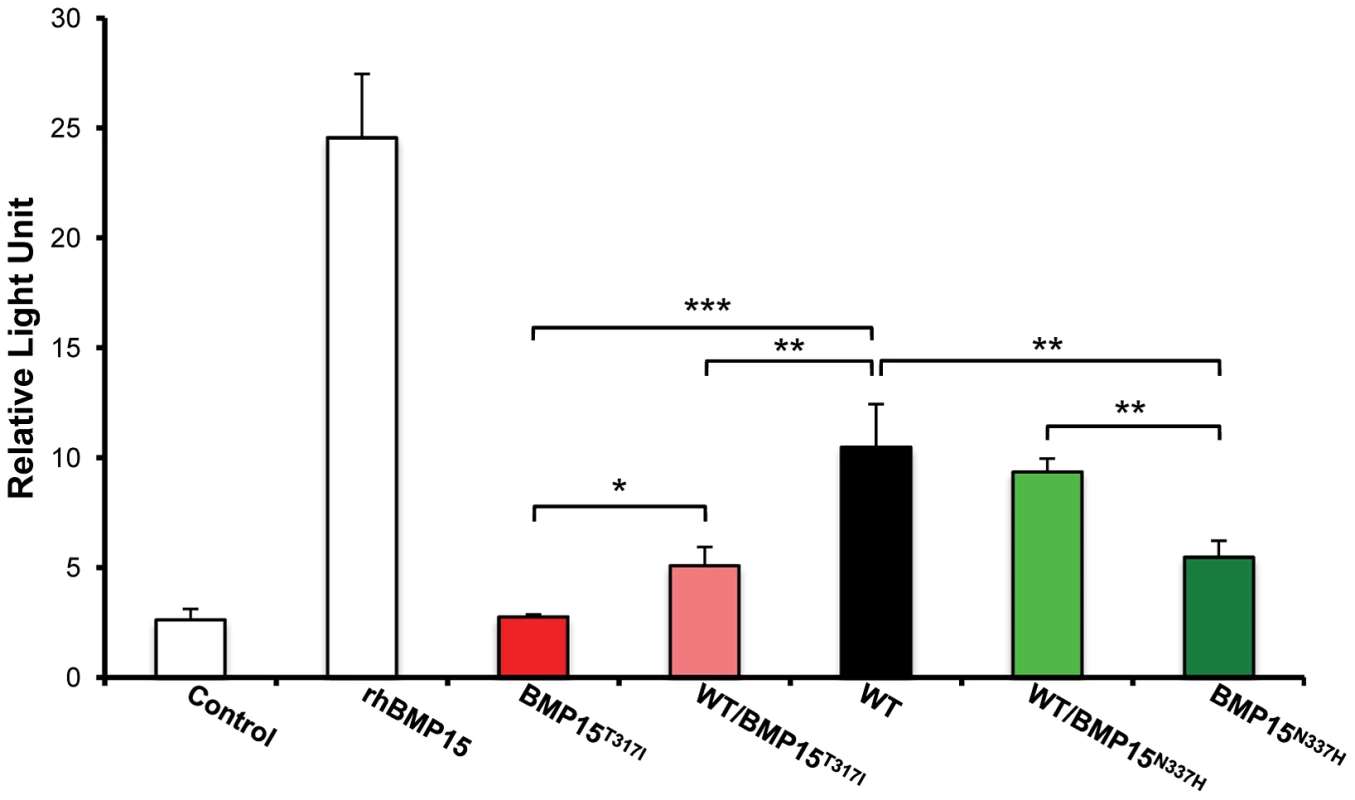
In vitro reporter luciferase assay from COV434 granulosa cells transiently transfected with empty vector +/− 100 ng of recombinant human BMP15 (Control +/− rhBMP15) or wild-type human BMP15 expressing vector (WT) or the 2 different BMP15 variant vectors (BMP15T317I (FecXGr); BMP15N337H (FecXO)) obtained by directed-mutagenesis. Results are expressed as Means±SD of the relative light unit (RLU) from 3 independent experiments in triplicate for each condition. Pairwise statistical comparisons using a one-way ANOVA test between means were performed and results of statistic test are symbolized by stars: * = p<5E−02; ** = p<1E−02 and *** = p<1E−03. Discussion
Some sheep breeds are naturally highly prolific and they are very informative for the studies of reproductive genetics and physiology. Genetic studies have indicated that LS and OR can be genetically determined by the action of single genes with a major effect, named fecundity (Fec) genes [38]. All the three Fec genes discovered so far belong to the TGFß superfamily: BMPR1B gene (FecB) located on OAR6 [18], [19], [20], GDF9 gene (FecG) located on OAR5 [14], [21], [22] and BMP15 gene (FecX) located on the X chromosome [13], [14], [15], [16], [17].
Effect of mutations on the prolificacy phenotype
Segregation of major genes increasing LS and OR was suspected in the French Grivette and the Polish Olkuska sheep flocks, respectively. In the Olkuska breed, a previous study had rejected the hypothesis of the segregation of 2 already known mutations, FecXI and FecBB [37]. Due to an atypical highly prolific phenotype with no sterile individual and no presence of streak ovaries in both breeds, a priori excluding the FecX gene, a GWAS based on a case/control design was performed to identify a causal locus in each population. A significant association was found for a cluster of SNPs located on the X chromosome close to the BMP15 gene suggesting its possible involvement. We identified 2 novel non-conservative mutations in the BMP15 gene called FecXGr and FecXO for the Grivette and the Olkuska variants, respectively. However, due to the specific genetic background of each breed as well as the difference of measured traits (OR vs LS), at this stage, the effect of the mutations appears to be slightly different for Olkuska and Grivette. Noteworthy, FecXGr and FecXO led to highly prolific ewes at the homozygous status in contradiction with other BMP15 variants described so far. Indeed, all the FecX variants (6 different mutated alleles) were associated with an increased LS/OR in heterozygous animals but sterility in homozygous animals [13], [14], [15], [16]. Then, it was unexpected to identify mutations in the BMP15 gene exclusively responsible for a high prolificacy phenotype. Nevertheless, an identical phenotypic effect has been recently shown for a new GDF9 allele called FecGE in the Brazilian Santa Ines sheep breed. Silva et al., [22] described for the first time, a polymorphism in the GDF9 gene that increases the ovulation rate (82%) and prolificacy (58%) in fertile homozygous ewes. Therefore, our two novel mutations (FecXGr and FecXO) as well as FecGE are associated with a prolific phenotype in homozygous ewes.
Effect of mutations on the intrinsic BMP15 protein properties
Both FecXGr and FecXO mutations are closely located into two very well conserved domains of the sheep, cow, pig, human and mouse BMP15 proteins. The BMP15 protein belonging to the TGFβ family is translated as a pre-proprotein which consists of a signal peptide, a large proregion and a mature peptide (reviewed in [39]). In sheep, 2 (FecXR [16], [17] and FecXG [14]) and 6 (FecXH [14], FecXI [13], FecXL [15], FecXB [14], FecXGr and FecXO) BMP15 mutations have been found in the proregion and the mature protein, respectively. The effect of sheep variants seems tightly related to the kind of mutations described. Indeed, 3 out of the 8 mutations identified so far are aminoacids deletion (FecXR [16], [17]) or premature stop codon (FecXG and FecXH) [14] in the BMP15 sequence impairing consequently the production of the BMP15 active form. Our mutations are located in the mature BMP15 protein, on both sides of the FecXL mutation. A putative misfolding inhibiting the maturation/production of the BMP15 protein was suspected in the case of the FecXL mutation which leads to a substitution of a cysteine with a tyrosine in one of the six conserved cysteines involved in the characteristic folding of the TGFβ factors [15]. The identified FecXGr and FecXO mutations convert threonine to isoleucine and asparagine to histidine in the French Grivette and the Polish Olkuska sheep populations, respectively. These two mutations clearly affect the intrinsic properties of the BMP15 protein since they correspond to substitutions of polar aminoacids by nonpolar and basic aminoacids suspected to modify consequently its three dimensional structure.
Effect of mutations on the BMP15 signaling pathway
Functionally, both FecXGr and FecXO mutations clearly impaired the BMP15 signaling as shown by our in vitro test in COV434 cells. Indeed, BMP15T317I induced a complete inhibition of BMP15 in vitro activity whereas the BMP15N337H only partially disturbed BMP15 signaling. The partial loss-of-function observed for the BMP15N337H mutation fits well with the hyperprolific phenotype in homozygous Olkuska sheep following the dosage-sensitive hypothesis of BMP15 action on OR [13], [40]. In vitro effect of FecXGr on the BMP15 pathway is similar to the FecXL variant [32] although prolificacy phenotypes of homozygous ewes are opposite i.e. FecXL/FecXL animals are sterile whereas FecXGr/FecXGr individuals are highly prolific. Such functional discrepancy has been already observed when comparing FecXL with other known non-conservative substitutions in the BMP15 mature peptide such as FecXI and FecXB. Indeed, functional analyses of FecXI and FecXB mutations indicated that neither V299D nor S367I substitutions are able to alter the production, processing, homodimerization, or biological activity of the BMP15 protein [41] while homozygous animals are sterile. The underlying mechanism occurs through the heterodimerization of BMP15 with the closely related oocyte-derived GDF9 protein [41], [42], [43] and their cooperative effect on granulosa cells [44], [45], [46]. Although the FecXGr mutation totally impaired the BMP15 direct signaling, it would still maintain a biological activity through its interaction with GDF9 suggesting a normal folding of the BMP15 protein in contrast to the FecXL variant [15]. Furthermore, we hypothesized that the BMP15T317I null mutation doesn't prevent the heterodimerization process with GDF9 compared to the FecXI variant for which the heterodimer signaling pathway has been implicated [41], [42], [43]. To better understand molecular effects of both FecXGr and FecXO mutations, independent actions of BMP15 and GDF9 as well as their synergic role remains to be determined.
Model of the ovulation quota control by the various Fecundity mutations
Based on the current knowledge, the mono ovulation quota seems to be tightly controlled by the triple action of i) BMP15 homodimer and its signaling pathway through BMPR1B, BMPR2 and SMAD1/5/8 [47], ii) BMP15/GDF9 heterodimer signaling also through SMAD2/3 [46] and iii) GDF9 homodimer signaling through TBR1, BMPR2 and SMAD2/3 [48], [49] (Figure 6A). However, the integration of these 3 pathways under a threshold activity precociously blocks the folliculogenesis and leads to sterility. This could be illustrated by the FecXL, H, G, R effect directly abolishing the BMP15 production [13], [14], [15], [16], [17] thus impairing at least BMP15 homodimer and BMP15/GDF9 heterodimer signaling. Moreover, when the FecXI or FecXB mutants are coexpressed with normal GDF9, the secretion of both BMP15 and GDF9 is significantly reduced [42]. Although effects of FecGH and FecGT variants have never been tested in vitro, it is strongly thought that GDF9 loss-of-function mutations might alter both GDF9 homodimer and BMP15/GDF9 heterodimer pathways leading to infertile animals. When the activity of the combined action of the 3 pathways is in between the sterility threshold and the normal level, i.e. heterozygous status, OR and LS are increased through modulation of granulosa cells proliferation and gonadotropin sensitivity [38], [40]. It is likely that Fecundity mutations resulting in hyperprolificacy without sterility might affect either totally the BMP15, GDF9 homodimers signaling pathways or partially the homodimer and heterodimer signaling pathways (Figure 6B and 6C). Our FecXGr and FecXO as well as FecGE mutations support this hypothesis. Indeed, the FecXO mutation would mainly have an effect on the BMP15/GDF9 heterodimer signaling via the heterodimerization process whereas the FecXGr mutation clearly impairs the BMP15 homodimer pathway through the BMP15 homodimerization or the binding to its receptor (Figure 6D). This assumption is close to the Booroola (FecBB) homozygous hyperprolific phenotype where a loss-of-function mutation altered the BMP15 receptor BMPR1B [50] and then suggested that only the BMP15 homodimer signaling pathway was affected. Concerning the FecXGr mutation for which the abolishment of the Smad 1/5/8 pathway was observed, it is also possible that another BMP15 signaling pathway, the TAK1 pathway, remains functional maintaining a biological BMP15 activity [46]. This assumption would explain why homozygous FecXGr/FecXGr ewes are not sterile.
Fig. 6. Hypothesized overview of ovulation quota control by the various Fecundity mutations. 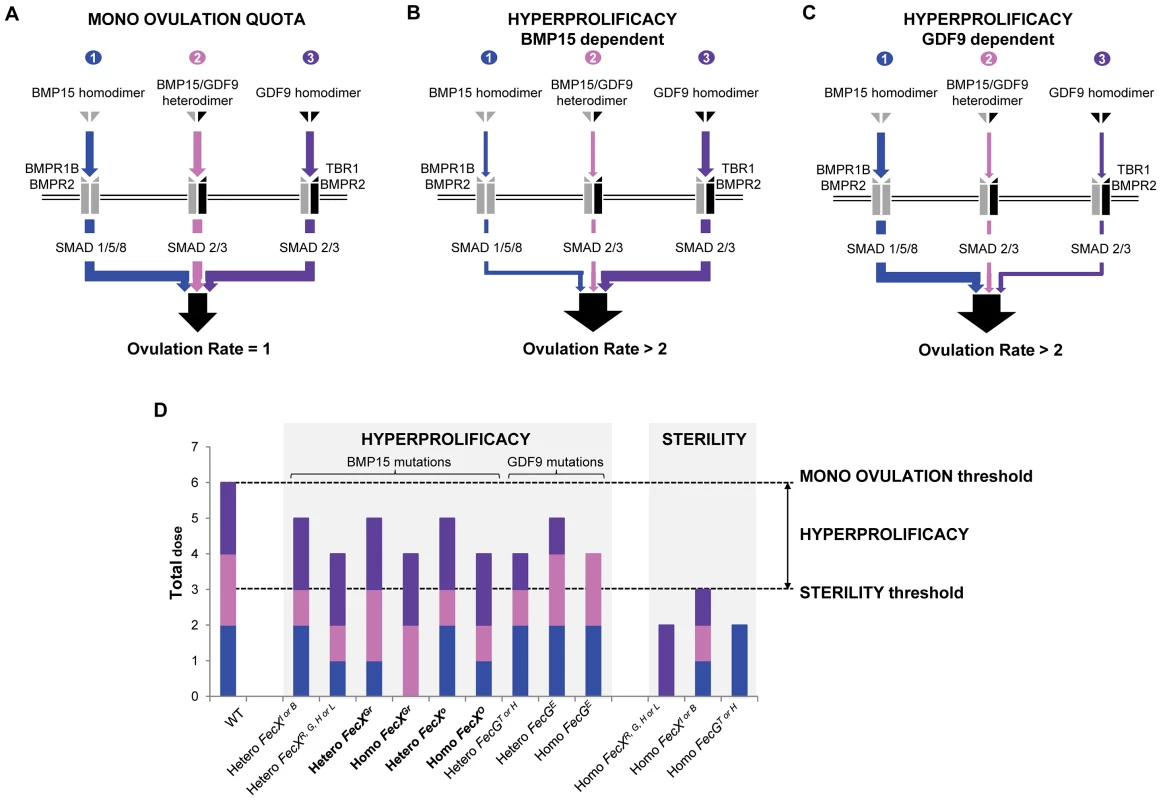
(A) The mono ovulation quota is tightly controlled by the integrative action of i) BMP15 homodimer through BMPR1B, BMPR2 and SMAD1/5/8, ii) BMP15/GDF9 heterodimer through SMAD2/3 and iii) GDF9 homodimer through TBR1, BMPR2 and SMAD2/3. (B) Increased of ovulation rate due to FecX mutations. This drawing represents an overview of hyperprolificacy dependent of BMP15 variants. Heterozygous or homozygous FecX mutations leading to an increased OR affect either the signaling pathways of BMP15 homodimer, BMP15/GDF9 heterodimer or both whereas the GDF9 homodimer signaling pathway remains stable. (C) Increased of ovulation rate due to FecG mutations. This drawing represents an overview of hyperprolificacy dependent of GDF9 variants. Heterozygous or homozygous FecG mutations leading to an increased OR impair either the signaling pathways of GDF9 homodimer, BMP15/GDF9 heterodimer or both whereas the BMP15 homodimer signaling pathway remains stable. (D) Dose sensitive effect of each FecX and FecG mutations on the BMP15, GDF9 homodimers and BMP15/GDF9 heterodimer signaling pathways. Based on the various in vitro tests performed, we assigned a dose (n = 0, 1 or 2) to each signaling pathway in an attempt to explain the prolificacy phenotype observed for all the FecX and FecG mutations. When the ovulation rate is normal ( = 1), i.e. the WT situation, 2 doses of BMP15 homodimer (blue), GDF9 homodimer (purple) and BMP15/GDF9 heterodimer (pink) were considered. As example, in the case of the FecXGr mutation where the BMP15 homodimer signaling pathway seemed clearly affected at homozygous and heterozygous status (as shown on Figure 5), we assumed that the BMP15/GDF9 heterodimer signaling pathway remained totally active. Comparison between human and sheep BMP15 mutations
The BMP15 protein has also been described in women to play critical roles in female fertility. Indeed, a large number of mutations in the BMP15 gene have been identified in women with POF [24],[29],[30],[31] and in patients with OHSS [33], [34] but none are in common between women and sheep. Interestingly, all the human BMP15 mutations described so far and involved in the POF syndrome were found at a heterozygous status. Therefore, heterozygous carriers exhibit an ovarian phenotype [29] similar to sterile homozygous FecX ewes which present an early folliculogenesis arrest, ovarian dysgenesis and streak ovaries [13], [15]. In contrary, heterozygous and homozygous ewes carrying FecXGr or FecXO mutations show an increase of LS and OR with normal and functional ovaries. Our data suggest that these two variants are novel kind of BMP15 mutations responsible for a new prolificacy phenotype as it was already shown in the case of the FecGE allele [22]. Noteworthy, BMP15 was recently involved in women suffering of OHSS since a BMP15 variant (BMP15−9G) was associated with this disorder [33], [34], similarly to reported effects of heterozygous BMP15 mutations in sheep. Although the connection between the BMP15−9G SNP and OHSS was mainly attributable to the considerably high number of heterozygous patients, a few women cases BMP15−9G/BMP15−9G homozygous were also identified and contribute to the significant signal obtained [33]. Therefore, the atypical phenotypic effect (high prolificacy) observed in ewes carrying FecXGr or FecXO mutations seems somewhat comparable to the OHSS in women at both heterozygous and homozygous status. Further studies need to be conducted to characterize the physiological ovarian phenotype of the highly prolific Grivette and Olkuska individuals.
In summary, our results bring new insights into the key role played by the BMP15 protein in ovarian function. The BMP15 gene is almost the only gene identified to date whose mutations in mammals as sheep and human result in impaired early folliculogenesis and POF as well as excessive ovulation rate and OHSS. Although the BMP15 biological effects might be specie-specific, parallels between phenotypes in women and sheep carrying BMP15 mutations emphasize the importance of the sheep model to further determine the involvement of BMP15 in ovarian physiology and pathophysiology in women.
Materials and Methods
Animals
The French Grivette population is a local breed mainly located in Rhône and Loire French departments. Ewes present naturally good maternal characteristics and a high prolificacy with more than 2 newborns per dam and per year. The very high variability of LS among related ewes suggested the segregation of a major gene affecting this trait. To test this assumption, 29 highly prolific ewes (LS≥2.7) considered as cases and 11 normally prolific ewes (LS≤1.8) considered as controls were selected to perform a genome-wide association study (GWAS) using a case/control design.
The Polish Olkuska population is a native Polish long-wool breed, traditionally kept in the southern region of the country, near Olkusz and Cracow. The Olkuska is an endangered prolific breed which in 90thies comprised only about 100 ewes dispatched in a few flocks, two of them belonging to Cracow and Warsaw Agricultural Universities that ensured the survival of this breed (reviewed in [36]). At present, due to a successful conservation program, the population increased to over 800 ewes. The main feature of the breed is its exceptional prolificacy, well over 200%, with also a very large variability of LS and OR for related animals. A wide range of mean individual OR (from 1.33 on 9 records to 6.20 on 15 records) led to assume the segregation of a putative major gene increasing OR. 29 cases ewes with high repeated OR (μ≥3.00) and 35 controls ewes with normal OR (μ≤2.00) were chosen to conduct a GWAS. Ovulation rates have been determined by laparoscopy. The laparoscopy was performed using a 5 mm diameter Vega S endoscope by experienced operators.
The description of the study design is presented in Table S1.
To confirm the effect of mutations on LS and OR in both Grivette and Olkuska populations, additional individuals have been genotyped and phenotyped. In total, 360 Grivette ewes coming from 9 flocks for which LS had been measured at least twice have been analyzed. In addition, laparoscopies performed at least 3 times have been obtained on 27 Grivette ewes in order to validate the effect of the mutation on OR. For the Olkuska population, the effect of the mutation on OR has been estimated from 103 ewes checked by laparoscopy (n>3) and LS data (n>2) have been retrieved for 137 animals.
Samples
Jugular venous blood samples were collected by venipuncture. Genomic DNA was extracted from blood samples following a salt-based DNA extraction as described in [15]. For the Grivette population, all procedures were approved by the “Direction Départementale des Services Vétérinaires de Haute-Garonne” (approval number C31-429-01) for the agricultural and scientific research agency INRA (French National Institute for Agricultural Research), and conducted in accordance with the Guide for the Care and Use of Agricultural Animals in Research and Teaching. For the Olkuska population, all procedures were performed with permission of the animal welfare commission from the University.
Genotyping analyses
Genotyping for the whole genome scan was performed on the Illumina OvineSNP50 Genotyping Beadchip according to the manufacturer's protocol (http://www.illumina.com). Individuals with a Call Rate <0.98 were excluded. SNPs were excluded if they showed a Call Freq <0.98, a minor allele frequency (MAF) <0.01 in cases or controls or significant deviation from Hardy-Weinberg equilibrium (HWE) in the controls (p<1E−06). Non polymorphic SNPs and markers with no position on the OARv2.0 map (http://www.livestockgenomics.csiro.au/sheep/) were also eliminated. Finally, an initial design of 54241 SNPs available on the Illumina OvineSNP50 Genotyping Beadchip and 104 individuals (40 and 64 ewes of the French Grivette and Polish Olkuska protocols respectively) was reduced to final datasets of 47290 SNPs analysed in 39 individuals for the French Grivette population and 46451 SNPs analysed in 63 individuals for the Polish Olkuska population for further analyses as showed in Table S1.
Genotyping of both FecXGr and FecXO mutations was performed by allele-specific amplification using the KASPar SNP genotyping system and followed by fluorescence detection on a ABI7900HT. KASPar assays were carried out in 5 µL reactions according to the KBioscience published conditions (http://www.kbioscience.co.uk/).
The primers used in this study are listed in Table S3.
Haplotype reconstruction analyses
The most likely linkage phase for each individual was determined using a coalescent theory approach under a Bayesian population genetic model implemented in the PHASE program [51], [52].
DNA sequencing analyses
Long-range PCR amplifications were performed using the Long PCR Enzyme Mix provided by Fermentas (http://www.fermentas.de) and internal primers were used for the sequencing reaction realized via the BigDye Terminator v3.1 Cycle Sequencing Kit (http://www.appliedbiosystems.com).
The primers used in this study are listed in Table S3.
Construction of BMP15 expression plasmids
BMP15 gene ovine variants (FecXGr/oBMP15T317I and FecXO/oBMP15N337H) were introduced by site-directed mutagenesis into the pCShBMP15wt vector, containing a full-length human BMP15 wild-type cDNA leading to the hBMP15T316I and hBMP15N336H, respectively. For the sake of simplicity, the numbering of ovine BMP15 was kept all along the manuscript. As previously described [32], mutagenesis reaction for each variant was performed using the QuickChange Site-Directed Mutagenesis kit (Stratagene) and specific couples of primers (Table S3).
Transient transfections and luciferase reporter assay
A COV434 human granulosa cells line stably expressing the BMP responsive element (BRE) - luciferase reporter was transiently transfected with BMP15 expressing vectors as already described by Rossetti et al. [32]. Briefly, COV434 cells were transfected in triplicate with 500 ng/well of pCShBMP15 vector expressing the wild-type or the mutant each alone or in equal combination (250 ng each) by using Fugene HD (Roche Applied Sciences, Indianapolis, IN). Cells were also transfected with the pCS2 empty vector and treated or not by 100 ng/ml of rhBMP-15 (R&D Systems, Minneapolis, MN), as positive or negative control, respectively. Forty hours after transfection, cells were lysed in Passive Lysis Buffer and assayed for luciferase activity using the Dual Luciferase reporter Assay kit (Promega). Luminescence in relative light units (RLU) was measured in a Fluoroskan Ascent instrument (Labsystems, Oy, Finland).
Statistical analyses
Single-marker association analyses were conducted using a Fisher's exact test and a Bonferroni correction has been applied to check for significance levels. The chromosome-wide and genome-wide values have been established as mentioned by Balding et al. [53]. Briefly, Bonferroni correction was applied to both the genome-wide and chromosome-wide analyses. The p-values generated were evaluated according to an adjusted significance threshold generated by dividing the 0.05 threshold by the total number of tests (number of SNPs considered) performed in each case (whole genome or whole chromosome). Statistical analyses were done using the PLINK software [54].
Haplotypic association analysis specifying all haplotypes in sliding windows of a fixed number of SNPs (n = 20) was also performed for X chromosome locus using chi-squared and logistic regression methods similar to other recent approaches [55], [56]. Empirical significance levels were calculated using the maximum statistic permutation approach (max(T), n = 1000).
For genotype effect on OR or LS and reporter luciferase assays, differences between means were analyzed by one-way ANOVA followed by Neuman-Keuls post-hoc test to compare between conditions. p<5E−02 was considered statistically significant. All the results were presented as means±SD.
Supporting Information
Zdroje
1. MatzukMM, BurnsKH, ViveirosMM, EppigJJ (2002) Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science 296 : 2178–2180.
2. GilchristRB, LaneM, ThompsonJG (2008) Oocyte-secreted factors: regulators of cumulus cell function and oocyte quality. Hum Reprod Update 14 : 159–177.
3. SuYQ, SugiuraK, EppigJJ (2009) Mouse oocyte control of granulosa cell development and function: paracrine regulation of cumulus cell metabolism. Semin Reprod Med 27 : 32–42.
4. DongJ, AlbertiniDF, NishimoriK, KumarTR, LuN, et al. (1996) Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature 383 : 531–535.
5. NilssonEE, SkinnerMK (2002) Growth and differentiation factor-9 stimulates progression of early primary but not primordial rat ovarian follicle development. Biol Reprod 67 : 1018–1024.
6. GilchristRB, RitterLJ, MyllymaaS, Kaivo-OjaN, DragovicRA, et al. (2006) Molecular basis of oocyte-paracrine signalling that promotes granulosa cell proliferation. J Cell Sci 119 : 3811–3821.
7. SpicerLJ, AadPY, AllenD, MazerbourgS, HsuehAJ (2006) Growth differentiation factor-9 has divergent effects on proliferation and steroidogenesis of bovine granulosa cells. J Endocrinol 189 : 329–339.
8. HusseinTS, FroilandDA, AmatoF, ThompsonJG, GilchristRB (2005) Oocytes prevent cumulus cell apoptosis by maintaining a morphogenic paracrine gradient of bone morphogenetic proteins. J Cell Sci 118 : 5257–5268.
9. OrisakaM, OrisakaS, JiangJY, CraigJ, WangY, et al. (2006) Growth differentiation factor 9 is antiapoptotic during follicular development from preantral to early antral stage. Mol Endocrinol 20 : 2456–2468.
10. JuengelJL, BodensteinerKJ, HeathDA, HudsonNL, MoellerCL, et al. (2004) Physiology of GDF9 and BMP15 signalling molecules. Anim Reprod Sci 82–83 : 447–460.
11. InagakiK, ShimasakiS (2010) Impaired production of BMP-15 and GDF-9 mature proteins derived from proproteins WITH mutations in the proregion. Mol Cell Endocrinol 328 : 1–7.
12. OtsukaF, McTavishKJ, ShimasakiS (2011) Integral role of GDF-9 and BMP-15 in ovarian function. Mol Reprod Dev 78 : 9–21.
13. GallowaySM, McNattyKP, CambridgeLM, LaitinenMP, JuengelJL, et al. (2000) Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat Genet 25 : 279–283.
14. HanrahanJP, GreganSM, MulsantP, MullenM, DavisGH, et al. (2004) Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries). Biol Reprod 70 : 900–909.
15. BodinL, Di PasqualeE, FabreS, BontouxM, MongetP, et al. (2007) A novel mutation in the bone morphogenetic protein 15 gene causing defective protein secretion is associated with both increased ovulation rate and sterility in Lacaune sheep. Endocrinology 148 : 393–400.
16. Martinez-RoyoA, JuradoJJ, SmuldersJP, MartiJI, AlabartJL, et al. (2008) A deletion in the bone morphogenetic protein 15 gene causes sterility and increased prolificacy in Rasa Aragonesa sheep. Anim Genet 39 : 294–297.
17. MonteagudoLV, PonzR, TejedorMT, LavinaA, SierraI (2009) A 17 bp deletion in the Bone Morphogenetic Protein 15 (BMP15) gene is associated to increased prolificacy in the Rasa Aragonesa sheep breed. Anim Reprod Sci 110 : 139–146.
18. MulsantP, LecerfF, FabreS, SchiblerL, MongetP, et al. (2001) Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. Proc Natl Acad Sci U S A 98 : 5104–5109.
19. SouzaCJ, MacDougallC, CampbellBK, McNeillyAS, BairdDT (2001) The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1 B (BMPR1B) gene. J Endocrinol 169: R1–6.
20. WilsonT, WuXY, JuengelJL, RossIK, LumsdenJM, et al. (2001) Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biol Reprod 64 : 1225–1235.
21. NicolL, BishopSC, Pong-WongR, BendixenC, HolmLE, et al. (2009) Homozygosity for a single base-pair mutation in the oocyte-specific GDF9 gene results in sterility in Thoka sheep. Reproduction 138 : 921–933.
22. SilvaBD, CastroEA, SouzaCJ, PaivaSR, SartoriR, et al. (2011) A new polymorphism in the Growth and Differentiation Factor 9 (GDF9) gene is associated with increased ovulation rate and prolificacy in homozygous sheep. Anim Genet 42 : 89–92.
23. DixitH, RaoLK, PadmalathaV, KanakavalliM, DeenadayalM, et al. (2005) Mutational screening of the coding region of growth differentiation factor 9 gene in Indian women with ovarian failure. Menopause 12 : 749–754.
24. LaissueP, Christin-MaitreS, TouraineP, KuttennF, RitvosO, et al. (2006) Mutations and sequence variants in GDF9 and BMP15 in patients with premature ovarian failure. Eur J Endocrinol 154 : 739–744.
25. KovanciE, RohozinskiJ, SimpsonJL, HeardMJ, BishopCE, et al. (2007) Growth differentiating factor-9 mutations may be associated with premature ovarian failure. Fertil Steril 87 : 143–146.
26. MontgomeryGW, ZhaoZZ, MarshAJ, MayneR, TreloarSA, et al. (2004) A deletion mutation in GDF9 in sisters with spontaneous DZ twins. Twin Res 7 : 548–555.
27. PalmerJS, ZhaoZZ, HoekstraC, HaywardNK, WebbPM, et al. (2006) Novel variants in growth differentiation factor 9 in mothers of dizygotic twins. J Clin Endocrinol Metab 91 : 4713–4716.
28. ZhaoH, QinY, KovanciE, SimpsonJL, ChenZJ, et al. (2007) Analyses of GDF9 mutation in 100 Chinese women with premature ovarian failure. Fertil Steril 88 : 1474–1476.
29. Di PasqualeE, Beck-PeccozP, PersaniL (2004) Hypergonadotropic ovarian failure associated with an inherited mutation of human bone morphogenetic protein-15 (BMP15) gene. Am J Hum Genet 75 : 106–111.
30. Di PasqualeE, RossettiR, MarozziA, BodegaB, BorgatoS, et al. (2006) Identification of new variants of human BMP15 gene in a large cohort of women with premature ovarian failure. J Clin Endocrinol Metab 91 : 1976–1979.
31. DixitH, RaoLK, PadmalathaVV, KanakavalliM, DeenadayalM, et al. (2006) Missense mutations in the BMP15 gene are associated with ovarian failure. Hum Genet 119 : 408–415.
32. RossettiR, Di PasqualeE, MarozziA, BioneS, TonioloD, et al. (2009) BMP15 mutations associated with primary ovarian insufficiency cause a defective production of bioactive protein. Hum Mutat 30 : 804–810.
33. HanevikHI, HilmarsenHT, SkjelbredCF, TanboT, KahnJA (2011) A single nucleotide polymorphism in BMP15 is associated with high response to ovarian stimulation. Reprod Biomed Online 23 : 97–104.
34. MoronFJ, de CastroF, RoyoJL, MontoroL, MiraE, et al. (2006) Bone morphogenetic protein 15 (BMP15) alleles predict over-response to recombinant follicle stimulation hormone and iatrogenic ovarian hyperstimulation syndrome (OHSS). Pharmacogenet Genomics 16 : 485–495.
35. YoshinoO, McMahonHE, SharmaS, ShimasakiS (2006) A unique preovulatory expression pattern plays a key role in the physiological functions of BMP-15 in the mouse. Proc Natl Acad Sci U S A 103 : 10678–10683.
36. DavisGH (2005) Major genes affecting ovulation rate in sheep. Genet Sel Evol 37 Suppl 1: S11–23.
37. DavisGH, GallowaySM, RossIK, GreganSM, WardJ, et al. (2002) DNA tests in prolific sheep from eight countries provide new evidence on origin of the Booroola (FecB) mutation. Biol Reprod 66 : 1869–1874.
38. VinetA, DrouilhetL, BodinL, MulsantP, FabreS, et al. (2012) Genetic control of multiple births in low ovulating mammalian species. Mamm Genome 23(11–12): 727–40.
39. PersaniL, RossettiR, CacciatoreC, FabreS (2011) Genetic defects of ovarian TGF-beta-like factors and premature ovarian failure. J Endocrinol Invest 34 : 244–251.
40. FabreS, PierreA, MulsantP, BodinL, Di PasqualeE, et al. (2006) Regulation of ovulation rate in mammals: contribution of sheep genetic models. Reprod Biol Endocrinol 4 : 20.
41. LiaoWX, MooreRK, ShimasakiS (2004) Functional and molecular characterization of naturally occurring mutations in the oocyte-secreted factors bone morphogenetic protein-15 and growth and differentiation factor-9. J Biol Chem 279 : 17391–17396.
42. LiaoWX, MooreRK, OtsukaF, ShimasakiS (2003) Effect of intracellular interactions on the processing and secretion of bone morphogenetic protein-15 (BMP-15) and growth and differentiation factor-9. Implication of the aberrant ovarian phenotype of BMP-15 mutant sheep. J Biol Chem 278 : 3713–3719.
43. McIntoshCJ, LunS, LawrenceS, WesternAH, McNattyKP, et al. (2008) The proregion of mouse BMP15 regulates the cooperative interactions of BMP15 and GDF9. Biol Reprod 79 : 889–896.
44. McNattyKP, JuengelJL, ReaderKL, LunS, MyllymaaS, et al. (2005) Bone morphogenetic protein 15 and growth differentiation factor 9 co-operate to regulate granulosa cell function. Reproduction 129 : 473–480.
45. EdwardsSJ, ReaderKL, LunS, WesternA, LawrenceS, et al. (2008) The cooperative effect of growth and differentiation factor-9 and bone morphogenetic protein (BMP)-15 on granulosa cell function is modulated primarily through BMP receptor II. Endocrinology 149 : 1026–1030.
46. ReaderKL, HeathDA, LunS, McIntoshCJ, WesternAH, et al. (2011) Signalling pathways involved in the cooperative effects of ovine and murine GDF9+BMP15-stimulated thymidine uptake by rat granulosa cells. Reproduction 142 : 123–131.
47. MooreRK, OtsukaF, ShimasakiS (2003) Molecular basis of bone morphogenetic protein-15 signaling in granulosa cells. J Biol Chem 278 : 304–310.
48. VittUA, MazerbourgS, KleinC, HsuehAJ (2002) Bone morphogenetic protein receptor type II is a receptor for growth differentiation factor-9. Biol Reprod 67 : 473–480.
49. MazerbourgS, KleinC, RohJ, Kaivo-OjaN, MottersheadDG, et al. (2004) Growth differentiation factor-9 signaling is mediated by the type I receptor, activin receptor-like kinase 5. Mol Endocrinol 18 : 653–665.
50. FabreS, PierreA, PisseletC, MulsantP, LecerfF, et al. (2003) The Booroola mutation in sheep is associated with an alteration of the bone morphogenetic protein receptor-IB functionality. J Endocrinol 177 : 435–444.
51. StephensM, SmithNJ, DonnellyP (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68 : 978–989.
52. StephensM, DonnellyP (2003) A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73 : 1162–1169.
53. BaldingDJ (2006) A tutorial on statistical methods for population association studies. Nat Rev Genet 7 : 781–791.
54. PurcellS, NealeB, Todd-BrownK, ThomasL, FerreiraMA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 : 559–575.
55. SchaidDJ, RowlandCM, TinesDE, JacobsonRM, PolandGA (2002) Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 70 : 425–434.
56. ZaykinDV, WestfallPH, YoungSS, KarnoubMA, WagnerMJ, et al. (2002) Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered 53 : 79–91.
57. KyteJ, DoolittleRF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157 : 105–132.
58. ZimmermanJM, EliezerN, SimhaR (1968) The characterization of amino acid sequences in proteins by statistical methods. J Theor Biol 21 : 170–201.
Štítky
Genetika Reprodukční medicína
Článek The G4 GenomeČlánek Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance inČlánek RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations inČlánek Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during DevelopmentČlánek Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic LethalityČlánek DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 4
-
Všechny články tohoto čísla
- Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
- A Big Catch for Germ Cell Tumour Research
- The Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
- A Nonsynonymous Polymorphism in as a Risk Factor for Human Unexplained Cardiac Arrest with Documented Ventricular Fibrillation
- The Hourglass and the Early Conservation Models—Co-Existing Patterns of Developmental Constraints in Vertebrates
- Smaug/SAMD4A Restores Translational Activity of CUGBP1 and Suppresses CUG-Induced Myopathy
- Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
- The G4 Genome
- Extensive Natural Epigenetic Variation at a Originated Gene
- Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases
- The Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
- TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
- Aconitase Causes Iron Toxicity in Mutants
- Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
- Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
- Sex-Differential Selection and the Evolution of X Inactivation Strategies
- Identification of a Tissue-Selective Heat Shock Response Regulatory Network
- Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity
- RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in
- Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
- Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
- ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair
- Iron-Responsive miR-485-3p Regulates Cellular Iron Homeostasis by Targeting Ferroportin
- Mutations in Predispose Zebrafish and Humans to Seminomas
- Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands
- Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during Development
- All SNPs Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated SNPs
- Functional 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- Genetic Requirements for Signaling from an Autoactive Plant NB-LRR Intracellular Innate Immune Receptor
- SNF5 Is an Essential Executor of Epigenetic Regulation during Differentiation
- Dialects of the DNA Uptake Sequence in
- Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate–Invertebrate Gap
- Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing
- High-Resolution Mapping of Spontaneous Mitotic Recombination Hotspots on the 1.1 Mb Arm of Yeast Chromosome IV
- Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
- Evolution after Introduction of a Novel Metabolic Pathway Consistently Leads to Restoration of Wild-Type Physiology
- Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
- Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
- Signatures of Diversifying Selection in European Pig Breeds
- The Chromosomal Passenger Protein Birc5b Organizes Microfilaments and Germ Plasm in the Zebrafish Embryo
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- Regulates Synaptic Development and Endocytosis by Suppressing Filamentous Actin Assembly
- Sensory Neuron-Derived Eph Regulates Glomerular Arbors and Modulatory Function of a Central Serotonergic Neuron
- Analysis of Rare, Exonic Variation amongst Subjects with Autism Spectrum Disorders and Population Controls
- Scavenger Receptors Mediate the Role of SUMO and Ftz-f1 in Steroidogenesis
- DNA Double-Strand Breaks Coupled with PARP1 and HNRNPA2B1 Binding Sites Flank Coordinately Expressed Domains in Human Chromosomes
- High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells
- Comparative Genomics of and the Bacterial Species Concept
- Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming
- Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
- The Genetic Correlation between Height and IQ: Shared Genes or Assortative Mating?
- Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
- An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
- The Genome Organization of Reflects Its Lifestyle
- Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood
- Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate
- Site-Specific Phosphorylation of the DNA Damage Response Mediator Rad9 by Cyclin-Dependent Kinases Regulates Activation of Checkpoint Kinase 1
- Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin
- Identification of , a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis
- Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
- DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
- Asynchronous Replication, Mono-Allelic Expression, and Long Range -Effects of
- Differential Association of the Conserved SUMO Ligase Zip3 with Meiotic Double-Strand Break Sites Reveals Regional Variations in the Outcome of Meiotic Recombination
- Focusing In on the Complex Genetics of Myopia
- Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans
- Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types
- Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD
- Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse
- Genetic and Genomic Architecture of the Evolution of Resistance to Antifungal Drug Combinations
- Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs
- Functional Dissection of the Condensin Subunit Cap-G Reveals Its Exclusive Association with Condensin I
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The G4 Genome
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



