-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaMreB-Dependent Inhibition of Cell Elongation during the Escape from Competence in
In bacterial cells, like in their eukaryotic counterparts, precise spatiotemporal localization of proteins is critical for their cellular function. This study shows that the expression and the localization of the bacterial actin-like MreB protein are growth phase-dependent. During exponential growth, we previously showed that MreB, together with other morphogenetic factors, forms discrete assemblies that move in a directed manner along peripheral tracks. Here, we demonstrate that in cells that develop genetic competence during stationary phase, transcription of mreB is specifically activated and MreB relocalizes to the cell poles. Our findings suggest a model in which MreB sequestration by the late competence protein ComGA prevents cell elongation during the escape from competence.
Published in the journal: . PLoS Genet 11(6): e32767. doi:10.1371/journal.pgen.1005299
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005299Summary
In bacterial cells, like in their eukaryotic counterparts, precise spatiotemporal localization of proteins is critical for their cellular function. This study shows that the expression and the localization of the bacterial actin-like MreB protein are growth phase-dependent. During exponential growth, we previously showed that MreB, together with other morphogenetic factors, forms discrete assemblies that move in a directed manner along peripheral tracks. Here, we demonstrate that in cells that develop genetic competence during stationary phase, transcription of mreB is specifically activated and MreB relocalizes to the cell poles. Our findings suggest a model in which MreB sequestration by the late competence protein ComGA prevents cell elongation during the escape from competence.
Introduction
In response to nutritional deprivation and high population density, the rod-shaped model Gram-positive bacterium Bacillus subtilis enters stationary phase and develops diverse environmental adaptations, namely competence for genetic transformation, sporulation, cannibalism or biofilm formation [1]. These developmental programs are exquisitely regulated in order to anticipate starvation and optimize the survival of at least a fraction of the population. During the development of these adaptations, cells initiate a large reorganization of gene expression [2,3], protein localization [4,5] and cell shape [5].
In the case of genetic competence, the central regulator ComK activates the expression of more than a hundred genes [2,6,7]. Competence development in B. subtilis is a well-known bistable system [1]. Only a small fraction of a population (2 to 10%) expresses the ComK-dependent genes, and thus the large majority of the population remains in the non-competent state [8,9]. Within the ComK regulon, twenty-eight genes are essential for genetic transformation [10], a process defined as the genetic alteration of a competent cell by incorporation of foreign DNA in its genome. The remaining genes upregulated in the presence of ComK may be involved in functions other than transformation. Accordingly, it was proposed to rename the ComK-determined physiological state the K-state, a more neutral term than genetic competence [2]. For instance, it has been shown that growth is inhibited during the escape from competence. When the environmental conditions improve (e.g. upon dilution into fresh medium), non-competent cells rapidly resume growth whereas competent cells remain in a growth-limited state during which both cell elongation and cell division remain inhibited for more than 90 minutes before they start to grow again [11,12]. This delay relative to non-competent cells is thought to constitute a tightly regulated checkpoint to allow the repair of the chromosome following homologous recombination of the transforming DNA, before replication initiation [11,12]. Growth inhibition during the escape from competence is controlled at two levels: cell elongation is inhibited through the late competence peripheral protein ComGA [11] and cell division is inhibited by ComGA and the highly conserved protein Maf [11,12]. The ComGA-mediated mechanism that inhibits cell elongation during outgrowth remains unknown. After exhibiting a diffuse localization in the cytoplasm, ComGA accumulates preferentially at polar clusters where it co-localizes with other competence proteins to form the transformation machinery [4,13]. Upon dilution into fresh medium, ComGA stays at the poles for 120 minutes before delocalizing, presumably through degradation or inactivation, ultimately reversing elongation inhibition [12].
Among the different classes of proteins regulating bacterial cell elongation, the bacterial actin-like MreB proteins have been the most studied over the past fifteen years. MreB proteins (Mre, for Murein cluster e) are essential for cell morphogenesis in most non-spherical bacteria [14,15]. In exponentially growing rod-shaped cells, MreB proteins localize in membrane-associated assemblies that rotate perpendicularly to the long axis of the cell [16–21]. These MreB structures are thought to control cell elongation by directing the assembly and movement of macromolecular complexes that effect synthesis of the sidewalls (cell cylinder) during growth [14,16,17].
In B. subtilis, sidewall elongation during vegetative growth is controlled by the redundant action of three MreB isoforms: MreB, Mbl and MreBH [22]. mreB and mbl are essential under normal growth conditions [23,24], while mreBH is essential only under certain adverse conditions [22,25]. The mreB gene is found in the third position of an operon composed of seven genes; immediately upstream the mreCD morphogenes and the minCD division-related genes, and downstream maf, involved in division inhibition during competence [12], and radC, of yet unknown function. It has been shown that several promoters are located within or upstream the mreB operon [12,26,27]. mbl is found immediately downstream spoIIID, a gene encoding a sporulation-specific transcriptional regulator [28] and usd, a gene located upstream spoIIID and necessary for its translation [29]. A sigma-E dependent promoter, activating the mbl expression during sporulation, is located upstream usd and spoIIID [26,30]. However, it has been shown that expression of mbl during vegetative growth is ensured by a sigma-A dependent promoter located between spoIIID and mbl [26]. Finally, mreBH forms an operon with a small gene of unknown function, ykpC [26]. Transcription of the mreBH operon is driven by the alternative sigma factor sigma-I, which is induced during heat shock [31]. The specific expression of the three mreB isoforms, from different promoters depending on different sigma factors, is in agreement with their partial functional redundancy upon various stress conditions [22].
Interestingly, mreB and mbl were identified as competence-induced genes in a transcriptomic study [2]. However, a detailed profile of expression of these two genes throughout growth and stationary phase remained to be characterized, and a possible role of MreB-like proteins in stationary phase adaptations was not investigated so far. Here, we report a new role associated to MreB during genetic competence in B. subtilis. We show that mreB (but not mbl) belongs to the ComK regulon, and that in competent cells MreB forms a complex with several competence proteins. Additionally, MreB co-localizes with ComGA in polar clusters. We finally show that ComGA-dependent growth inhibition displayed by cells escaping the K-state also involves MreB. We propose a model in which ComGA sequesters MreB in order to prevent cell elongation during outgrowth and therefore the escape from competence.
Results
mreB is a competence-induced gene, regulated by ComK
In previous transcriptional profiling studies of B. subtilis grown to competence, all the genes of the mreB operon were found to be down-regulated in comK mutant cells relative to wild-type cells [2]. mbl was also down-regulated but only when mecA, which codes for the adaptor protein that targets ComK for proteolysis [32], was knocked-out to increase the percentage of competent cells [33]. It was proposed that expression of both mreB and mbl was ComK-dependent and thus induced during competence, although it could not be excluded that mbl expression was affected by ComK only in the pleiotropic mecA background [2].
We examined whether transcription of mreB, mbl and/or mreBH was specifically induced during competence. Fragments of different sizes (500 to 2300 bp) upstream the open-reading frames of mreB (Fig 1A), mbl (S2A Fig) and mreBH (S2B Fig) containing several promoters were fused to the firefly luciferase (luc) coding sequence. In the case of mreB, three promoters were previously identified: P1, upstream the maf-radC-mreBCD-minCD operon [12,26]; P2, inside maf [27] and P3, between radC and mreB [26,27] (Fig 1A). P1 and P3 are dependent on the major housekeeping sigma factor sigma-A, while P2 is dependent on extracytoplasmic sigma factors [26,27,34]. P1 also contains ComK binding boxes (Fig 1A) and was shown to drive expression of maf during competence [12]. We measured the transcription rate from three fragments upstream mreB: PmreB123, containing the three promoters; PmreB23, containing promoters P2 and P3, and PmreB3, containing P3 (Fig 1A) during growth (measured by OD600, S1 Fig) in competence medium (CM). During exponential growth, expression of luc fused to PmreB123, PmreB23 and PmreB3 was virtually identical (Fig 1B). The transcription rate progressively increased to reach a maximum during the transition from exponential growth to stationary phase, which marks the beginning of competence (T0). This indicated that in exponentially growing cells expression of mreB comes from P3. No transcript generated from P1 and P2 could be detected, even if P1 has been shown to drive the low, basal expression of maf during exponential growth [12]. Upon entering stationary phase (after T0), the transcription rate from all three fragments rapidly decreased (Fig 1B). Transcription from PmreB23 (P2 + P3) and PmreB3 (P3 alone) exhibited a relatively sharp and progressive decrease, reaching a low basal level approximately 3 h after T0. However, expression from PmreB123 (P1 + P2 + P3) was significantly higher and exhibited a prominent burst about 2 hours after T0 (T2, which corresponds to the time of maximal competence [35]). Thus, in stationary phase there was a substantial (4–6 fold, see inset in Fig 1B) increase in mreB transcription that came from P1, the promoter in front of the operon. This was consistent with the recent finding that maf is expressed during competence from P1, regulated by the master regulator ComK [12]. As expected, when we monitored mreB transcription in a comK mutant, the transcriptional burst observed at T2 was abolished and the transcription rate from PmreB123 was comparable to that from the two shorter fragments PmreB23 and PmreB3 (Fig 1C).
Fig. 1. mreB is a competence-regulated gene. 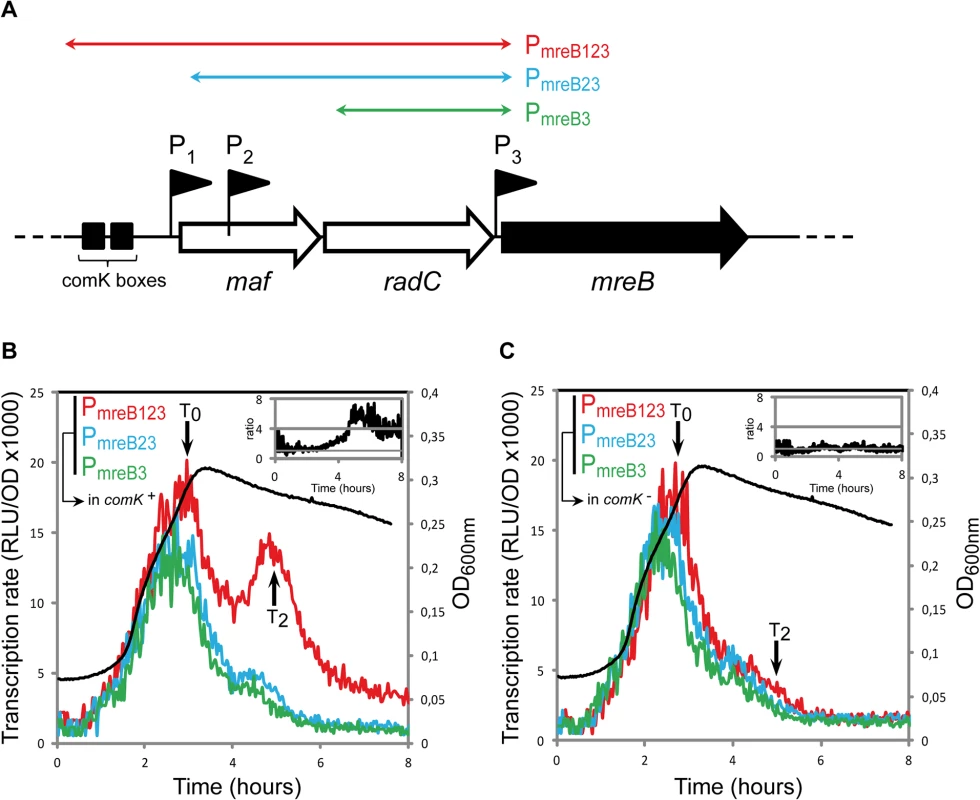
A. Partial map of the mreB operon. The three first genes of the operon, maf, radC and mreB are displayed. The three promoter’s upstream mreB (P1, P2 and P3) are represented by black flags. The two black boxes in front of the operon indicate the ComK binding sites that allow competence-specific over-expression of the operon [12]. The three colored double-headed arrows delimit the fragments used in the luciferase assay to characterize the expression coming from each promoter. PmreB123 (in red, strain NC91) contains the three promoters and the ComK boxes, PmreB23 (in blue, NC92) contains promoters P2 and P3, and PmreB3 (in green, NC93) only contains the last promoter in front of mreB, P3. B. Transcription profiles of strains expressing PmreB123-luc (in red, NC91), PmreB23-luc (in blue, NC92) or PmreB3-luc (in green, NC93) in a comK+ background during growth in competence medium at 37°C. Transcription rates are presented as the evolution of relative luminescence units corrected for OD (RLU/OD, left y-axis). As a reference, the NC91 growth curve (in black) is represented (OD600nm, right y-axis). The black arrows denote time points (in hours) relative to the beginning of competence (T0). The three cultures grew almost identically (see S1A Fig) The inset on the top right corner shows the evolution of the ratio between the expression detected from PmreB123 and PmreB3. C. Same as B except that transcription profiles were measured in a comK mutant background (in red, strain NC146; in blue, NC147; in green, NC148). As a reference, the NC146 growth curve (in black) is represented (OD600nm, right y-axis). The three cultures grew almost identically (see S1B Fig). The inset on the top right corner shows the evolution of the ratio between the expression detected from PmreB123 and PmreB3 during time in the absence of comK. Two promoters were previously identified for mbl: P1, a sigma-E-dependent promoter located upstream the usd gene and P2, a sigma-A-dependent promoter right upstream mbl (S2A Fig) [26,30,36]. Like mreB, mbl was transcribed predominantly during exponential growth, and maximum of expression was reached right before T0 (S2C Fig). Expression of mbl in exponentially growing cells came exclusively from P2. In contrast to mreB, however, expression of mbl was not reactivated in stationary phase and was not affected by ComK (S2C Fig), even though a small peak can be observed around T2. mbl was previously reported to be over-expressed in comK mutant cells only when mecA was also knocked-out [2]. Since mecA mutants are very pleiotropic [37,38], our results indicate that activation of mbl transcription in the comK- mecA- background was indirect, resulting from secondary effects of the absence of mecA. For mreBH, only a sigma-I-dependent promoter, induced during heat shock, has been identified [31]. Consistently, no transcription of mreBH was detected during growth in competence medium (S2D Fig).
Taken together, our findings indicate that mreB, but not mbl and mreBH, is a competence-induced gene, regulated by ComK.
MreB is associated to ComGA in competent cells
To provide insight into a possible role of MreB in competent cells, we sought to identify MreB binding partners during competence. To this end, MreB was fused to the sequential peptide affinity (SPA) tag [39]. Unlike cells lacking MreB, cells containing spa-mreB as only copy of mreB in their genome displayed normal morphology in both exponential and stationary phase (S3 Fig) indicating that the SPA-MreB fusion was functional. The strain expressing the SPA-MreB fusion was grown to T2 in CM at 37°C, and MreB-associated proteins were purified and identified by mass spectrometry. Strains expressing no SPA-tagged protein and a SPA fusion to PerR, a non-related protein of B. subtilis, were used as negative controls. Interestingly, several competence proteins (ComGA, Maf, ComEB, ComC and ComFA) were specifically and reproductively detected in the MreB pull-down complexes (Table 1). Among these proteins, ComGA was the most abundant in the complex based on the Protein Abundance Index (PAI, established according to [40]). ComGA was co-purified with SPA-MreB well above the contaminant value found in the control strains (Table 1), indicating that their co-purification was specific. ComEB, ComC and ComFA were specifically co-purified, and Maf was greatly enriched in the SPA-MreB eluate relative to the control strains (Table 1). Taken together, these results indicated that MreB is associated with several competence proteins in B. subtilis.
Tab. 1. MreB is in complex with competence proteins. 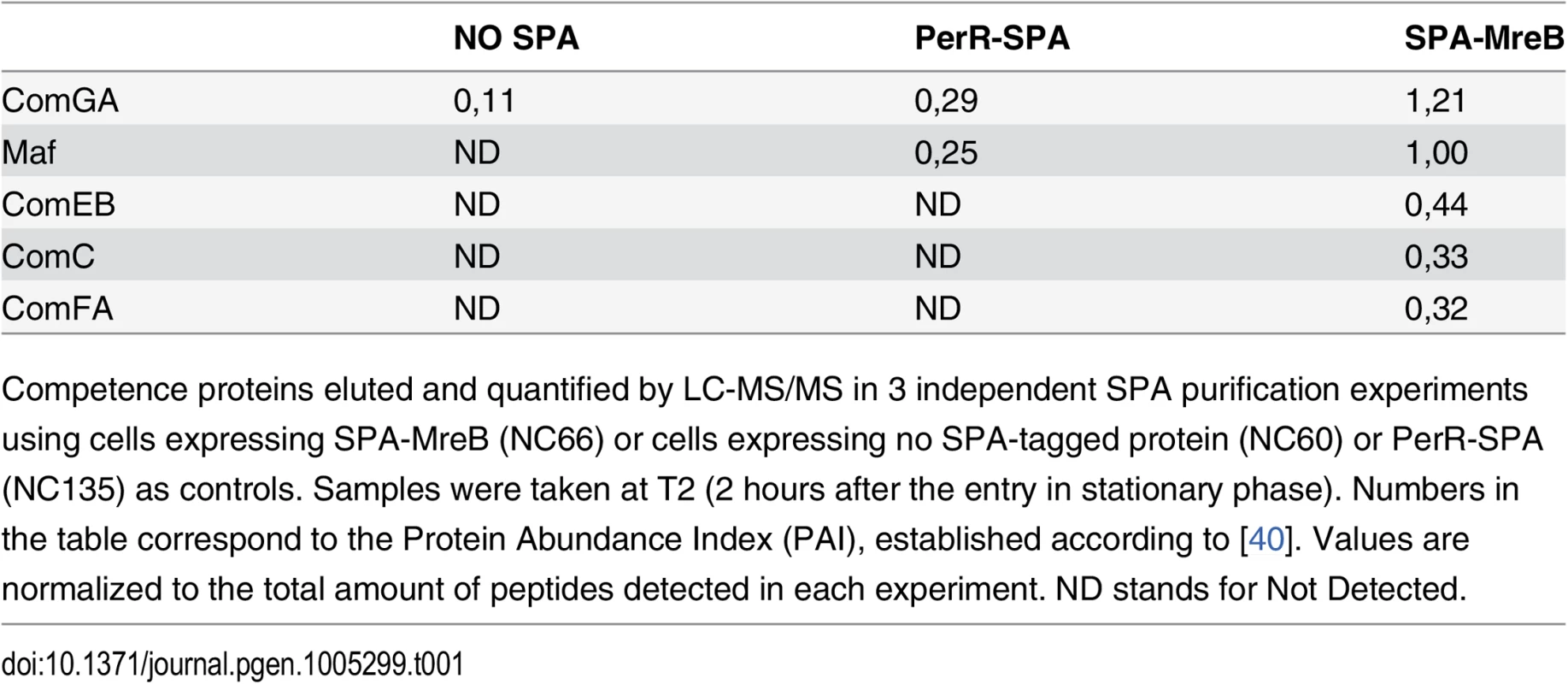
Competence proteins eluted and quantified by LC-MS/MS in 3 independent SPA purification experiments using cells expressing SPA-MreB (NC66) or cells expressing no SPA-tagged protein (NC60) or PerR-SPA (NC135) as controls. Samples were taken at T2 (2 hours after the entry in stationary phase). Numbers in the table correspond to the Protein Abundance Index (PAI), established according to [40]. Values are normalized to the total amount of peptides detected in each experiment. ND stands for Not Detected. Next, we determined whether MreB displays a specific localization in the subpopulation of competent cells. We have shown that expression of mreB is complex, driven from three different promoters (Fig 1). To avoid a possible artifact of overexpression and/or misregulation, we replaced mreB by gfp-mreB at the native locus expressed under control of the native mreB regulatory sequences (Pnativegfp-mreB), without leaving any scar or resistance cassette in the vicinity (see Methods for details, S4A Fig and S1 Movie). We then analyzed nativeGFP-MreB localization in cells that natively expressed a functional ComGA-RFP fusion (PnativecomGA-rfp) as a marker for competence. Strikingly, in stationary phase cells (T2), most nativeGFP-MreB signal disappeared from the membrane and became diffuse in the cytoplasm (‘b’ cells in Figs 2A and S4C). In competence-expressing cells at T2, in addition of exhibiting a diffuse signal, nativeGFP-MreB formed clusters at one or both cell poles (‘a’ cells in Figs 2A and S4C). All polar MreB clusters (n > 200) were found to co-localize with ComGA polar clusters, while no nativeGFP-MreB signal was found in 15% (n > 200) of ComGA polar assemblies (Figs 2A and S4C). In the absence of comGA, MreB polar clusters were never observed and nativeGFP-MreB fluorescence signal was diffuse in all cells (n > 200) (Fig 2B). Control experiments showed that this co-localization was not due to bleed-through of the bright ComGA-RFP signal into the GFP channel (S4E Fig). In a given field of view, the integrated fluorescence signal of nativeGFP-MreB per cell was more than 3 times higher in competent (AU = 67.2 ±17.6, n = 102) than in non-competent (AU = 20.8 ±11.6, n = 103) cells, indicating that ComK-dependent expression of mreB (Fig 1) leads to increased levels of MreB protein in competent cells.
Fig. 2. MreB co-localizes with ComGA polar clusters in competent cells. 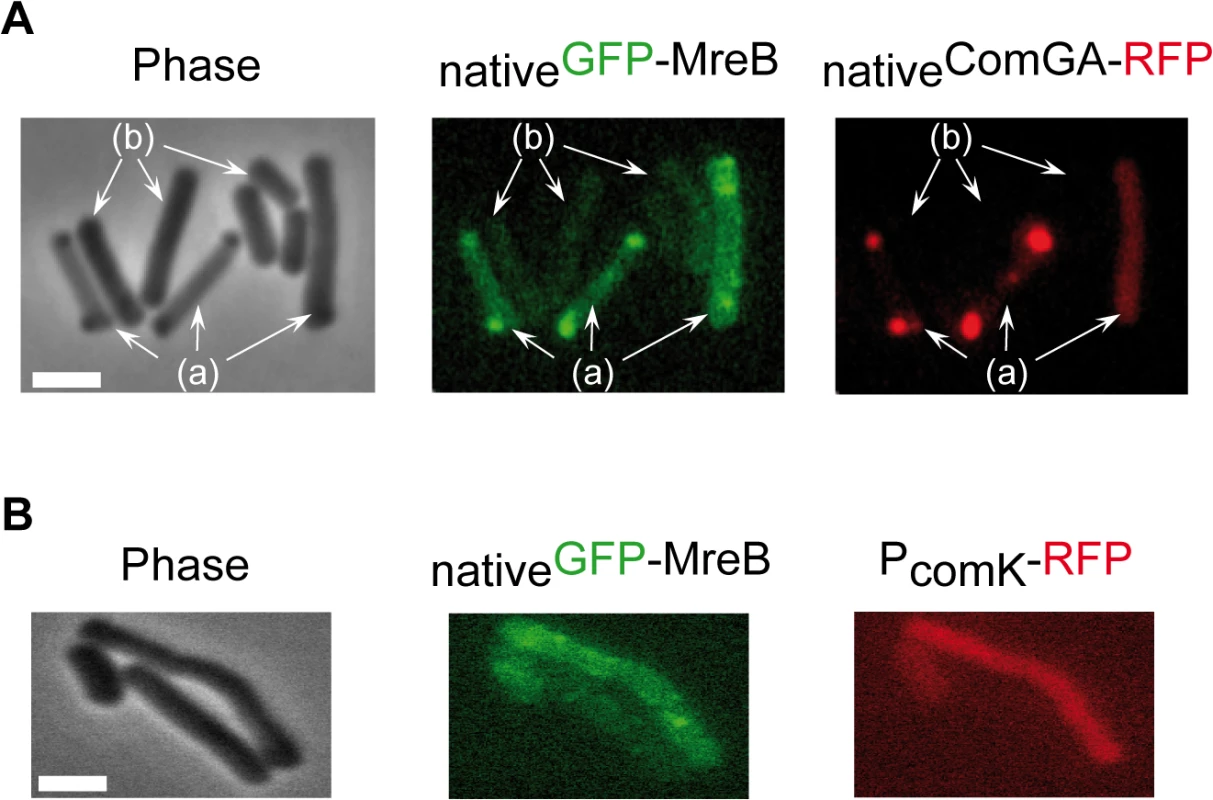
A. Co-localization of nativeGFP-MreB and nativeComGA-RFP (strain NC121) in competent (displaying a RFP signal, a) and non-competent cells (b) of B. subtilis. Cells were grown at 37°C in competence medium to T2 and imaged by conventional epifluorescence microscopy. Typical group of cells imaged by (left to right): Phase contrast (Phase); GFP (green) and RFP (red) channels. The co-localization experiment of nativeMbl-GFP and nativeComGA-RFP is shown in S4D Fig as control. Scale bar, 1µm. B. Localization of nativeGFP-MreB in the comGA mutant background (strain NC215) at T2. The competent cells are easily identifiable as they express RFP under the control of the comK promoter. Scale bar, 1µm. In exponential growth, a functional nativeMbl-GFP fusion displayed the characteristic ‘motile patches’ localization (S4B Fig and S2 Movie). However, in contrast to nativeGFP-MreB, at T2 a nativeMbl-GFP fusion was still localized in membrane-associated patches (albeit no longer motile, S4 Movie) along the sidewalls, which did not co-localize with ComGA polar clusters (S4D Fig). Thus, in competent cells MreB, but not Mbl, relocalizes into polar clusters that colocalize with, and are dependent on, the multi-functional competence protein ComGA.
MreB is not required for genetic transformation
ComGA was first described for its essential role in natural genetic transformation [2]. We then tested if MreB could play a role during this process. Strikingly, transformation efficiency of in-frame mreB null mutants was increased about a hundred fold relative to the wild-type strain, while mbl and mreBH mutants had transformation efficiencies comparable to that of the wild-type (Table 2). However, in cells lacking mreB, both the percentage of competent cells and the timing of competence development were not affected (S5A and S6A Figs respectively).
Tab. 2. Transformation efficiencies. 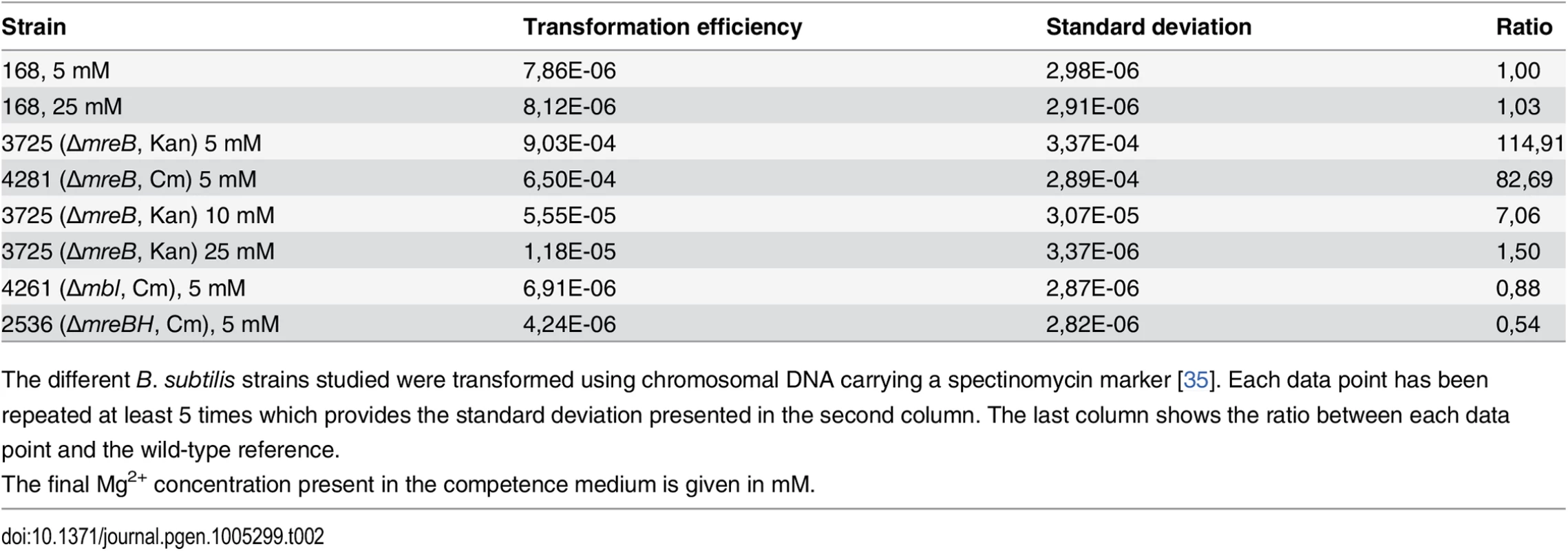
The different B. subtilis strains studied were transformed using chromosomal DNA carrying a spectinomycin marker [35]. Each data point has been repeated at least 5 times which provides the standard deviation presented in the second column. The last column shows the ratio between each data point and the wild-type reference. High concentrations of magnesium (Mg2+) rescue the viability and shape defects of mreBs and other mutants involved in different aspects of cell wall synthesis by a yet unknown mechanism [14]. It has been proposed that Mg2+ may stiffen the cell wall, compensating for structural defects associated to the absence of mreB [41]. CM is traditionally supplemented with 5 mM Mg2+ [35]. Remarkably, increasing Mg2+ concentrations in CM progressively rescued the mreB transformation phenotype (Table 2). At 25 mM Mg2+, the transformation efficiency of mreB mutant cells was down to wild-type levels (Table 2). Taken together, these results suggested that the effect of MreB in transformation is indirect. They also raised the interesting possibility that specific cell wall defects could promote transformation in B. subtilis. One hypothesis was that the assembly or localization of the transformation apparatus across the cell wall was affected in the absence of mreB. To investigate this, we compared the localization of the transformation machinery in the wild-type and mreB mutant backgrounds using our nativeComGA-RFP fusion. The dynamic localization of ComGA during competence has been extensively described [4]. In wild-type cells developing competence ComGA first appears diffuse in the cytoplasm (S7B Fig). Then, ComGA forms clusters associated to the inner face of the membrane with an important bias for the regions near the poles (S7C and S7D Figs), where it co-localizes with other main competence proteins to form the transformation machinery [4]. The number of ComGA focus per wild-type competent cell varies from one to nine, but the large majority of wild-type competent cells (41%, n>1500) display a single ComGA polar cluster (S5C and S5D Figs). The percentage of competent cells that displayed nativeComGA-RFP clusters at T2 (S5B Fig) and among these the number of ComGA clusters (S5C Fig) were significantly higher in cells lacking mreB relative to wild-type cells. More specifically, the majority of mreB mutant competent cells (37%, n>1500) displayed three foci (S5C and S5E Figs). Expression of the comGA gene (S6B and S6D Figs) and ComGA protein levels (S6E Fig) were nevertheless unaffected in the mreB mutant. At high Mg2+ concentrations, mirroring the recovery of wild-type transformation efficiency of the mreB mutant, the distribution of the number of ComGA clusters per competent mreB mutant cell shifted back to wild-type levels (S5B Fig). We concluded that MreB is not directly required for natural transformation and that ComGA localization might be impacted by the cell wall integrity.
ComGA inhibits cell elongation through MreB to prevent the escape from competence
It was shown that ComGA is also required for inhibition of cell elongation in cells exiting competence [11], while MreB directs cell elongation in exponentially growing cells [14,16,17]. We hypothesized that MreB could be involved in inhibition of cell elongation during competence escape through its association with ComGA. To test this, we performed outgrowth experiments using a ComK-GFP construct to distinguish competent from non-competent cells, as previously described [11]. Wild-type, ΔcomGA, ΔmreB and ΔmreB ΔcomGA mutant strains were grown to T2, when maximal competence is achieved, and diluted 20-fold into fresh medium. Samples were taken prior to dilution (T2) and 90 minutes after dilution (T2+90) for size and morphology characterization. At T2 competent and non-competent cells were indistinguishable in length for all strains (S3 Table) [12]. At T2+90, non-competent cells had resumed growth and division [11,12]. Competent cells of the wild-type strain (Fig 3A) were only slightly longer than at T2 (S3 Table), confirming the previously reported growth limitation imposed during the escape from competence [11]. ΔmreB competent cells also remained in a growth-limited state after 90 minutes of outgrowth and were significantly shorter than wild-type competent cells (Fig 3C and 3F and S3 Table) as previously reported for exponentially growing mreB mutant cells [41]. In contrast, ΔcomGA competent cells were filamentous and often bent (Fig 3B and 3F and S3 Table), indicating that ComGA directly or indirectly inhibits cell elongation during the escape of competence [11]. However, when mreB was knocked out in the ΔcomGA mutant strain, the filamentous phenotype of ΔcomGA competent cells was rescued, and the average cell length of the ΔmreB ΔcomGA mutant was similar to that of wild-type competent cells at T2+90 (Fig 3D and 3F and S3 Table).
Fig. 3. ComGA inhibits elongation during the escape from competence through MreB. 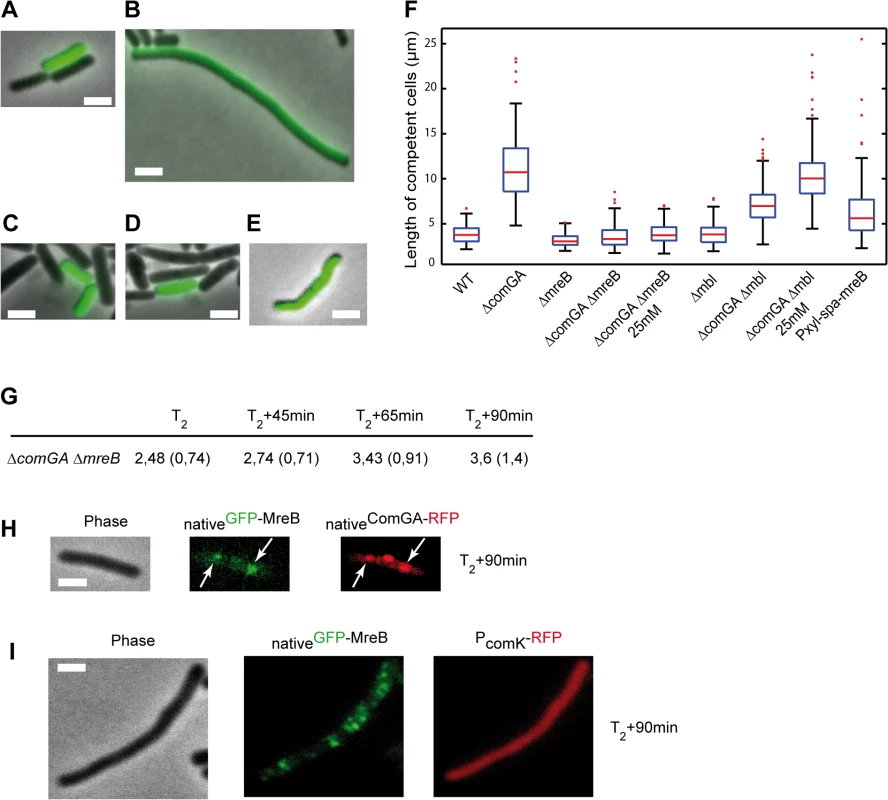
A-E. Representative examples of the morphology of competent cells of the wild-type (NC59, A), the ΔcomGA (NC164, B), the ΔmreB (NC161, C), the ΔmreB ΔcomGA (NC169, D) and the Pxyl-spa-mreB (NC197, E) strains 90 minutes after dilution into fresh competence medium (T2+90). Competent cells can be identified as they display nativeComK-GFP signal. Scale bars, 2µm. F. Boxplots of the length of competent cells of the wild-type (NC59), ΔcomGA (NC164), ΔmreB (NC161), ΔmreB ΔcomGA (NC169), Δmbl (NC162), Δmbl ΔcomGA (NC170) and Pxyl-spa-mreB (NC197) strains at T2+90 during the outgrowth experiment. Cells of the ΔmreB ΔcomGA (NC169) and Δmbl ΔcomGA (NC170) strains were grown and diluted in normal competence medium (5 mM Mg2+) and in competence medium with a final Mg2+ concentration of 25 mM. At least 100 cells were counted for each strain and each condition. Details about this statistical analysis and the way this graph was constructed are presented in the Methods section. G. Mean cell length of competent cells of the ΔmreB ΔcomGA strain (NC169) during the outgrowth experiment. Samples were taken for cell length measurements prior to dilution (T2) and 45, 65 and 90 minutes after dilution into fresh competence medium. At least 100 cells were measured for each time point. Standard deviations are indicated between brackets. H. Co-localization by epifluorescence microscopy of nativeGFP-MreB and nativeComGA-RFP (NC121) after dilution and 90 minutes of growth (T2+90min). Left to right: Phase contrast (Phase); GFP (green) and RFP (red) channels. White arrows indicate the co-localizing MreB and ComGA clusters. Scale bar, 1 µm. I. Localization of nativeGFP-MreB in the absence of comGA during outgrowth. Cells expressing nativeGFP-MreB and PcomK-RFP (as a marker of competence) in the comGA mutant background (strain NC215) were imaged by epifluorescence microscopy at T2+90min. Scale bar, 1µm. Wild-type rod shape is restored in mreB and mbl null mutants by addition of 25 mM Mg2+ to the growth medium, while addition of 2.5 mM Mg2+ is sufficient to restore wild-type growth rate of mreB mutants [41,42]. Consistently, ΔmreB mutant cells were viable and displayed wild-type growth and moderate cell shape defects in classic CM (i.e. 5mM Mg2+) (S8 Fig and S5 Movie). To exclude an indirect effect due to the inability of mreB-like mutants to elongate properly at low Mg2+ concentrations, we repeated the outgrowth experiments in CM containing 25 mM Mg2+. At T2+90, the length of ΔcomGA ΔmreB competent cells was similar in conventional CM (5 mM Mg2+) and in CM with 25 mM Mg2+ (Fig 3F and S3 Table). In contrast, deletion of mbl ameliorated but did not rescue the ΔcomGA filamentous phenotype, and in the presence of 25 mM Mg2+ ΔcomGA Δmbl competent cells filamented like ΔcomGA competent cells (Fig 3F). Taken together, these results indicated that MreB plays a direct role in the growth limitation imposed during the escape from competence. However, it was still plausible that in the absence of MreB, competent ΔcomGA cells did filament but started dividing during the 90 minutes of outgrowth, as previously shown for Δmaf ΔcomGA mutant cells [12]. If this was true, the average length of ΔmreB ΔcomGA double mutant cells would first increase (during filamentation) and then decrease (upon initiation of cell division) between T2 and T2+90. Measurement of the length of competent cells at different times during the outgrowth experiment showed that ΔmreB ΔcomGA cells length slightly but progressively increased their average length from T2 to T2+90 (Fig 3G), excluding that they had filamented and then divided.
These findings suggested that ComGA-mediated inhibition of cell elongation during the escape from competence also involves MreB. One possibility is that ComGA directly or indirectly sequesters MreB in competent cells to delay the initiation of cell elongation upon outgrowth. According to this prediction, over-production of MreB could totally or partially bypass the ComGA checkpoint and thus promote elongation of competent cells during outgrowth. Consistently, when native levels of MreB were increased by expressing the functional spa-mreB fusion (S3 Fig) in the presence of the endogenous copy of mreB, competent cells filamented in a manner similar to ΔcomGA cells after 90 minutes of outgrowth. The mean length of cells overproducing SPA-MreB was almost twice the mean length of wild-type cells and cell length distribution was much broader, with cells exceeding 20 µm in length (Fig 3F).
ComGA inhibits re-localization of MreB to the sidewalls during outgrowth
We found that at the time of maximum competence (T2) MreB forms polar clusters that co-localize with ComGA polar clusters and are dependent on the presence of ComGA (Fig 2). In addition, our findings suggest that MreB is involved, alongside ComGA, in the inhibition of cell elongation during outgrowth. We then verified if the localization of MreB and ComGA was still correlated during the escape from competence. After 90 minutes of outgrowth, MreB polar clusters were still present and co-localized with ComGA clusters in wild-type competent cells (Fig 3H). However, in the filamentous ΔcomGA competent cells, MreB had already re-localized into motile patches along the sidewalls (Fig 3I). These results suggested a direct correlation between ComGA-dependent polar localization of MreB and the absence of elongation during the escape from competence.
Expression of comGA in exponentially growing cells affects cell growth and morphology
Our findings above suggest a model in which ComGA would directly or indirectly sequester MreB in competent cells. Unfortunately, difficulties to purify active recombinant MreB proteins currently unable biochemical work with MreB proteins of B. subtilis [14] and thus the direct interaction between MreB and ComGA could not be tested in vitro. No direct protein-protein interaction between MreB and ComGA was detected in pairwise yeast two-hybrid assays using full-length proteins (S9 Fig). False negatives are nevertheless frequent in two-hybrid assays [43], and thus the absence of interaction in yeast did not exclude a true protein interaction. Alternatively, we analyzed the effect of expression of comGA during exponential growth. In wild-type cells, comGA is exclusively expressed during competence [12]. In the same background, unnatural expression of comGA during exponential phase from an inducible promoter, was reported to have no effect on growth [11]. However, we reasoned that defects due to the sequestration of MreB by ComGA could be masked by the partial functional overlap between the three MreB isoforms [22]. Thus, we analyzed the effect of over-expression of comGA from the very strong (although poorly repressed) hyperspank promoter (Phs) in both wild-type and mbl mutant cells growing in rich (LB) medium. The mbl mutant strain grew almost like the wild-type strain in LB (Fig 4A), and a low percentage of cells (11%, n = 400 at OD = 0.15) displayed mild morphological defects (arrows in Fig 4D). Expression of comGA in the wild-type background had virtually no effect on growth (Fig 4A) [11] and morphology (Fig 4B and 4C). However, growth of the mbl mutant carrying the Phs-comGA-rfp construct, was significantly affected both in the absence and (to a bigger extent) in the presence of inducer (Fig 4A). This result clearly indicated that over-expression of comGA is toxic in the absence of Mbl. Furthermore, the majority (65%, n = 400 at OD = 0.15) of comGA-overexpressing mbl mutant cells showed progressive bulging and aberrant morphologies including Y-shaped cells and polar bulges characteristic of mreB (but not mbl) mutant cells [22,44] (Fig 4E), indicating impairment of cell morphogenesis and explaining the lethal effects on growth. We concluded that when comGA is expressed in exponentially growing cells, MreB cannot fully compensate for the absence of Mbl. These findings were consistent with the hypothesis that ComGA sequesters MreB to prevent cell elongation and limit growth. We could not test the effect of expression of comGA on the localization of MreB in the mbl mutant background because GFP fusions to MreB do not support growth in a Δmbl ΔmreB background.
Fig. 4. Effect of comGA expression during exponential phase. 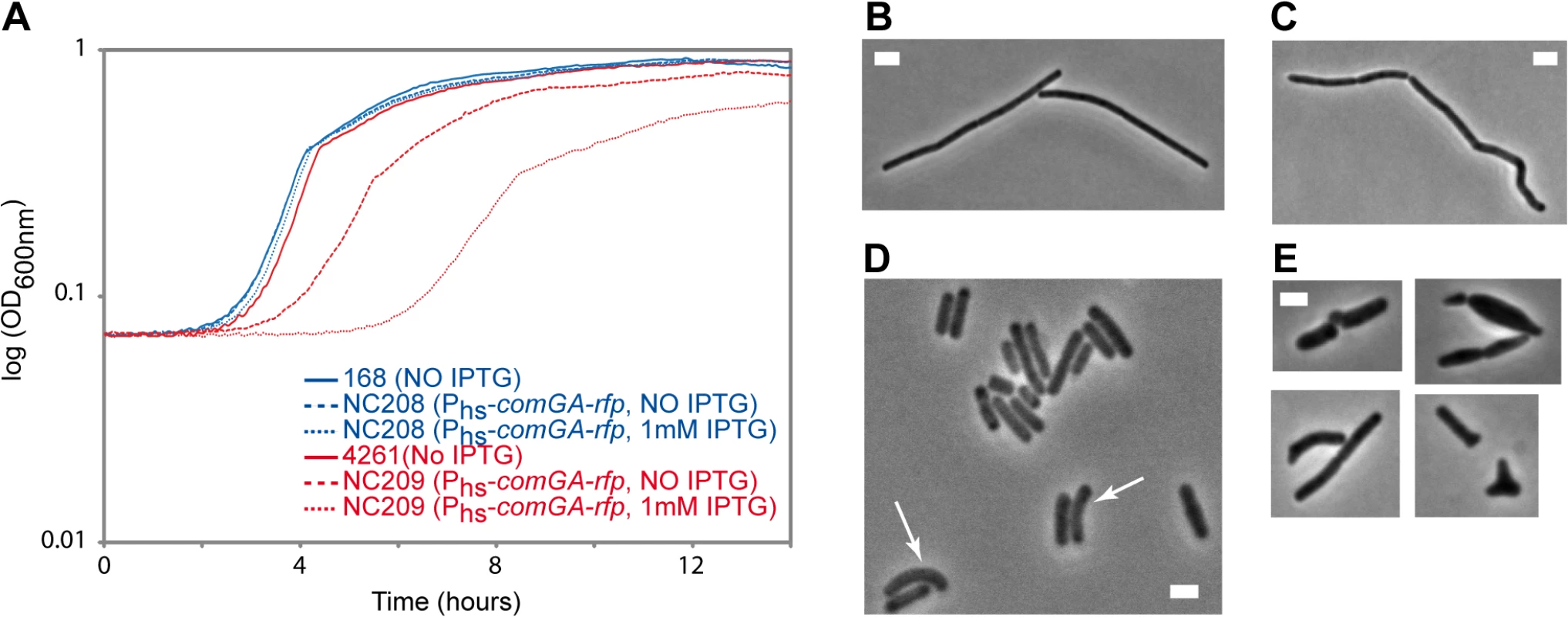
Growth and morphology of cells expressing comGA from the very strong (although poorly repressed) hyperspank promoter (Phs) in the wild-type and mbl mutant backgrounds. Cells were grown in LB medium at 37°C in the absence or in the presence of IPTG. A. Growth curves of the Phs-comGA-rfp (NC208, blue) and the Δmbl Phs-comGA-rfp (NC209, red) strains grown in the absence (NO IPTG, dashed lines) or in the presence (1mM IPTG, dotted lines) of inducer relative to the growth of their parental strains, 168 and 4261 (blue and red plain curves respectively). B-E. Phase contrast images of representative cells of the Phs-comGA-rfp (208, C) and Δmbl Phs-comGA-rfp (NC209, E) strains in the presence of 1 mM IPTG and their corresponding parental strains (B and D respectively) grown to OD600 = 0,15. Arrows in D point to mbl mutant cells that present morphological defects. Note that in D, the Δmbl cells are in average shorter than wild-type cells (B). Scale bar, 2µm. Discussion
Expression and localization of MreB are regulated in stationary phase and during competence
In bacterial cells, like in their eukaryotic counterparts, proteins localize to specific locations, often in a dynamic manner, during growth. Spatiotemporal localization of proteins is critical for their function and orchestrates cellular processes. In exponentially growing B. subtilis cells, the mreB gene is highly expressed and MreB assembles into membrane-associated patches that move processively around the cell to control sidewall elongation [16–20]. Here, we show that when B. subtilis cells enter stationary phase in competence medium, expression of mreB drastically decreases and MreB delocalizes from the membrane exhibiting a largely diffuse localization in the cytoplasm. Such transcriptional regulation of mreB and the disassembly of MreB patches from the membrane may inhibit deposition of peptidoglycan along the sidewalls during stationary phase. Additionally, we show that expression of mreB is reactivated in cells that develop competence. In competent cells, MreB relocalizes in polar clusters together with the late competence protein ComGA. Co-localization of MreB and ComGA at the cell poles persists for at least 90 minutes of outgrowth into fresh media. MreB subsequently relocalizes as motile patches along the sidewalls to reinitiate elongation. Altogether, these findings underline the importance of dynamic regulation of gene expression and protein localization for bacteria to adapt to changing environmental conditions.
Localization of the transformation apparatus is affected by the integrity of the cell wall
In cells lacking mreB, transformation efficiency was increased a hundredfold and the number of membrane-associated ComGA clusters was significantly higher than in wild-type cells. Both phenotypes were however rescued by high Mg2+ concentrations, suggesting that (i) MreB is not directly required for natural transformation in B. subtilis, and (ii) assembly of the transformation apparatus might be affected by structural features of the cell wall, as Mg2+ has been proposed to rigidify weakened cell-walls [41]. The transformation apparatus, which includes a type IV pilus-like structure that traverses the thick cell wall and is required for binding and importing the transforming DNA [45], preferentially localizes near the poles at the junction between the cylinder and the polar caps [4]. This region represents the interface between the sidewalls, which are intensively reshaped during growth, and the almost inert cell wall at the poles. Interestingly, this region is also chosen by phage SPP1 to bind and inject its DNA into the cytoplasm of B. subtilis [46]. During infection, SPP1 has to irreversibly bind to its receptor, YueB, encoded by a putative type VII secretion system gene cluster in B. subtilis [47,48]. YueB extends across the cell wall and also localizes at the junction between the cylinder and the polar caps [46]. Thus, this structurally differentiated region of the cell wall may contain positional information for the assembly of structures that need to cross the cell envelope. Initial assembly of the transformation apparatus pilus-like structure at these sites could then direct the localization of cytoplasmic competence-induced proteins such as ComGA at the inner leaflet of the cytoplasmic membrane. Specific defects in the structure or the organization of the cell wall of mreB mutant cells may favor the assembly of additional transformation apparatus at ectopic sites. Consistently, it has been shown that the absence of mreB induced the apparition of multiple sites containing polar material in E. coli cells [49,50]. Furthermore, inactivation of MreB in Pseudomonas aeruginosa led to the mislocalization of a normally polar type IV pilus [51].
A model for sequestration of MreB by ComGA to prevent cell elongation and delay the escape from competence
We show here that in competent cells mreB is specifically transcribed from the same promoter than maf and that MreB protein levels are increased relative to non-competent cells. Competent comGA mutant cells filament upon dilution into fresh medium [11]. These long comGA mutant cells are unable to divide because Maf is still present and inhibits cell division [12]. When mreB was deleted in a ΔcomGA background, competent cells did not filament during the early stages of competence escape. When mbl was deleted, the average length of ΔcomGA cells exiting competence was also slightly reduced. High Mg2+ concentrations fully rescued ΔcomGA cells elongation in the absence of mbl but not in the absence of mreB. Taken together, these findings indicate that elongation of cells escaping competence primarily depends on MreB and cannot be rescued by the redundant action of Mbl and/or MreBH. Mbl could nevertheless play a mild secondary role in this process. Consistently, a low level of transcription of mbl was detected during stationary phase at T2, while expression of mreBH was completely switched off. When mreB was overexpressed in a wild-type background, cells escaping competence exhibited a filamentous phenotype, like ΔcomGA cells. We hypothesize that in this condition excess of MreB can bypass the ComGA-mediated inhibition of elongation and activate cell wall synthesis. Finally, confirming the implication of the two proteins in order to limit cell elongation, MreB was found (i) in the same complex than several competence proteins and (ii) co-localizing with ComGA polar clusters at T2 and throughout the 90 minutes following dilution into fresh medium.
In the light of our results, we propose the model presented in Fig 5, in which ComGA inhibits cell elongation during the escape from competence by sequestering MreB, either directly or indirectly. No direct interaction between MreB and ComGA was detected in yeast two-hybrid assays and such interaction cannot be tested in vitro because active recombinant MreB of B. subitlis is currently not available for biochemical work [14]. However, we show here that expression of comGA in exponentially growing mbl mutant cells induces growth and morphological defects similar to those of mreB mutants. This result suggests that ComGA may be able to sequester MreB during exponential phase too. Therefore, if ComGA and MreB do not interact directly, then the potential protein(s) mediating their interaction during competence is (are) also expressed during vegetative growth. However, to date, all proteins found to co-localize with ComGA at the poles of competent cells are specifically over-produced during competence [4,52]. How the ComGA-MreB interaction is mediated remains an important question for future work.
Fig. 5. A model for sequestration of MreB by ComGA to prevent cell elongation and delay the escape from competence. 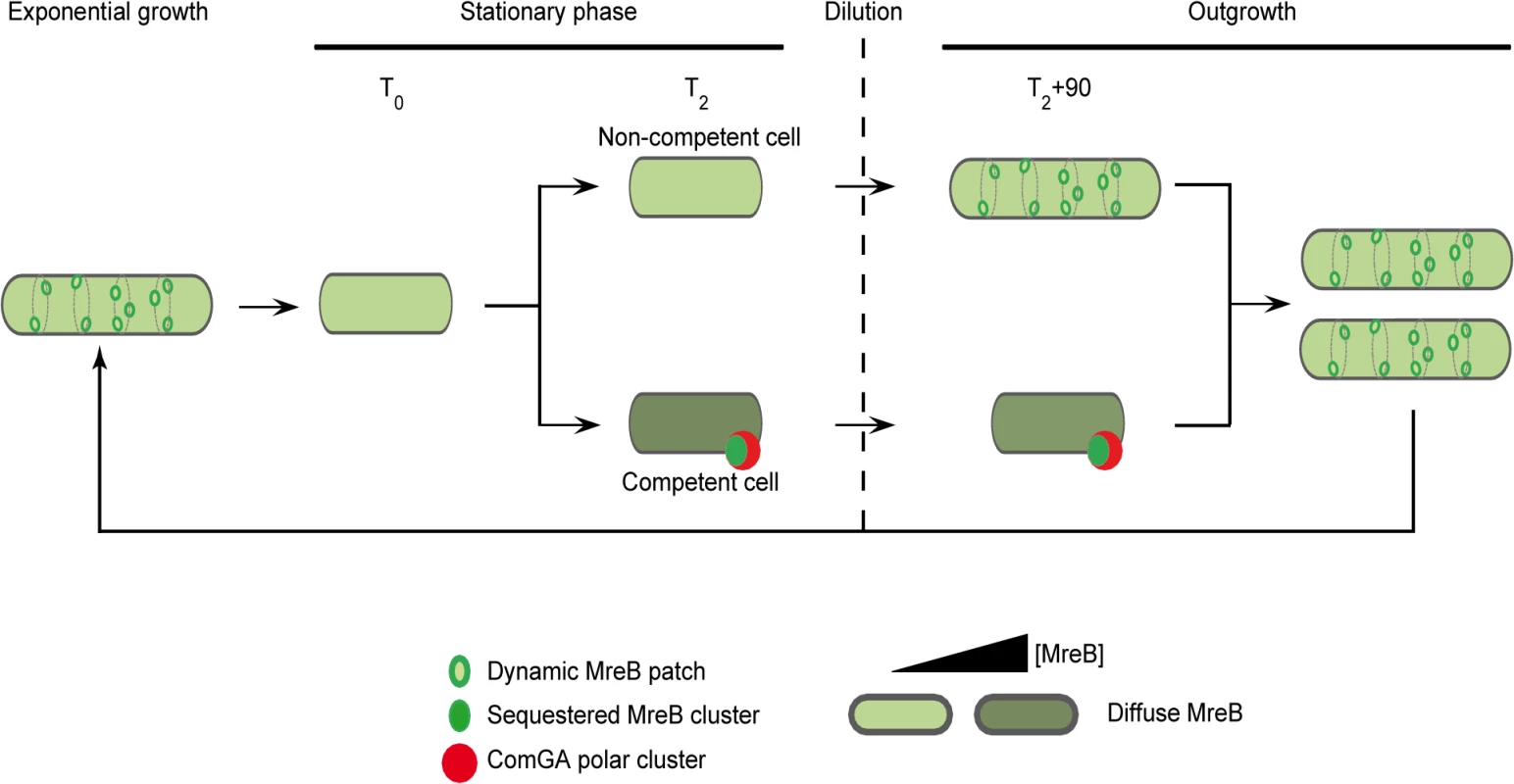
In exponentially growing cells, MreB forms patches associated to the inner face of the lateral membrane in order to direct cell wall synthesis. As nutrient concentration decreases in the environment cells enter stationary phase (T0). Meanwhile, MreB proteins delocalize from the membrane to the cytoplasm of all the cells. Cell wall synthesis is therefore limited and the cells are in average shorter than during exponential growth. Throughout the two first hours following T0, competence develops in a low percentage of cells (2 to 10% in a wild-type background). When the maximum of competence is reached (T2), MreB concentration has increased in the competent cells. Moreover, in addition of being diffuse in the cytoplasm, MreB co-localizes with ComGA at the competent cells poles. When this stationary phase culture (T2) is diluted into fresh medium (outgrowth), the non-competent cells quickly start to elongate and divide as MreB re-associates with the sidewalls. At the opposite, competent cells growth is limited for more than 90 minutes as MreB is sequestered by ComGA at the poles. We finally hypothesize that as the ComGA lock is released (after 2 hours of outgrowth), MreB relocalizes at the membrane and allow cell elongation. We propose that a higher MreB concentration allows the competent cells to grow faster while exiting competence compensating for the delay previously imposed. ComGA, a new regulator of the actin-like protein MreB?
General principles governing protein localization include capture by a cellular factor (e.g. interacting protein, DNA binding site, membrane domain or substrate) and self-assembly, where polymerization/depolymerization dictate the location of a protein at a given time [53]. Polymerization may also be regulated by binding proteins, like in the case of eukaryotic actin, where a myriad of actin-binding proteins (ABPs, [54,55]) regulate actin activity and dynamics. ABPs can nucleate, cross-link, bundle, anchor and regulate the state of polymerization of polymeric, filamentous actin (F-actin), and they can cap and stabilize the monomeric, globular actin (G-actin) pool in the cytoplasm.
Here, we show that in cells entering stationary phase, MreB dissociates from the sidewalls and becomes diffuse in the cytoplasm. Interestingly, it has been recently shown that the concentration of lipid-linked peptidoglycan precursors regulates the association of MreB to the membrane [56]. When precursors are depleted, MreB filaments disassemble into the cytoplasm. During the entry into stationary phase peptidoglycan precursor depletion probably occurs [57], as the metabolism slows down and the need of cell wall synthesis decreases, potentially explaining MreB relocalization. However, the details of the mechanism regulating the dynamic localization of MreB remain unknown. Numerous studies have identified a number of proteins that modulate FtsZ ring formation in B. subtilis [58–62] while the first ABP-like protein regulating MreB has yet to been found. It is plausible that one or several ABP-like protein(s), sensing the peptidoglycan precursor’s availability, promote MreB depolymerisation and/or stabilize the monomeric form of MreB in the cytoplasm.
In addition, we propose a model in which ComGA would sequester MreB in competent cells to prevent its localization to the sidewalls and therefore cell elongation. ComGA could then be considered as a new cellular regulator of the actin-like protein MreB. Only one protein that spatially regulates the MreB proteins has been reported in bacteria [63]. Indeed, the progressive depletion of RodZ leads to the misassembly of MreB into non-spiral structures before inducing a total loss of shape in Escherichia coli [63]. While RodZ can be considered as a positive regulator favoring the assembly of MreB at the right sites, ComGA could be classified as a negative regulator preventing the canonical MreB localization along cylindrical sidewalls.
The specific increase of the amount of MreB in competent cells might compensate for the delay imposed by its sequestration
We suggest that sequestration by ComGA spatially regulates MreB during competence in B. subtilis. When ComGA is eventually degraded or inactivated allowing competent cells to resume growth [11], excess of MreB relative to non-competent cells would be free to rapidly form membrane-associated patches and initiate fast elongation. ComK-dependent induction of mreB expression during competence would therefore compensate for the timing disadvantage imposed by genetic transformation. The two levels of regulation (i.e. gene expression and protein localization) might generate and orchestrate the pathway controlling simultaneously a delay in growth and a way to compensate for it.
One MreB isoform for one environmental adaptation?
It has been shown that MreB, Mbl and MreBH display partial functional redundancy in B. subtilis [22]. Overexpression of any one of the isoforms is sufficient to sustain lateral peptidoglycan synthesis and maintain cell shape in normal growth conditions. However, no single MreB isoform could support growth in various stress conditions, suggesting that multiplicity of MreB isoforms may become essential in specific environmental conditions [22]. Here we show that unlike mreB, mbl and mreBH are not specifically expressed during genetic competence. Consistently mbl and mreBH mutants displayed no competence-associated phenotypes. This specialization of MreB in competence further suggests that each isoform could be essential for specific environmental adaptations. A sigma-E sporulation specific promoter has been detected upstream mbl [26,30,36], while mreBH is part of the SigI regulon induced during heat stress [31,64]. Similarly to MreB in the context of competence, the localization and/or activity of Mbl and MreBH could be modulated by a regulator specifically produced during their respective adaptation. Future studies will reveal whether Mbl plays a role in sporulation and MreBH in stress response.
Methods
Microbiological methods
Bacillus subtilis strains were constructed by natural genetic transformation with selection for the appropriate antibiotic resistance marker. For transformation, competent cultures were prepared and incubated in competence medium (CM) with transforming DNA (~1 µg/ml) for 30 minutes at 37°C [35]. When needed, B. subtilis chromosomal DNA was prepared as detailed in [65]. Transformants were selected using 100 µg/ml spectinomycin, 10 µg/ml kanamycin, 5 µg/ml chloramphenicol, 16 µg/ml phleomycin and 1 µg/ml erythromycin. All the plates used to select transformants contained 25 mM of Mg2+. The details of all the new constructs in this publication are presented below. All new constructs were sequenced after introduction in the B. subtilis chromosome. B. Subtilis strains were grown in CM or LB media. When needed, the CM Mg2+ final concentration was increased to 25 mM. Strains are listed in S1 Table.
Construction of promoter-luciferase fusion strains
Because some of our genes of interest are in the middle of operons, we decided to clone our constructs (promoter + RBS + luciferase) at the ectopic amyE locus.
Fragments of different lengths upstream the genes of interest (mreB, mbl and mreBH) and ending right before the genes RBS were amplified by PCR from the B. subtilis chromosome. To amplify the fragments PmreB123, PmreB23, PmreB3 (Fig 1A), Pmbl12, Pmbl2 (S2A Fig) and PmreBH1 (S2B Fig) we used the primers MCS-PmreB1-F and RBS-PmreB-R, MCS-PmreB2-F and RBS-PmreB-R, MCS-PmreB3-F and RBS-PmreB-R, MCS-Pmbl1-F and RBS-Pmbl-R, MCS-Pmbl2-F and RBS-Pmbl-R and MCS-PmreBH1-F and RBS-PmreBH-R respectively. In parallel, we amplified by PCR the upstream (amy-Front and choramphenicol cassette) and downstream (amy-Back and luciferase gene) amyE fragments from the plasmid pUC18cm-luc [66] using primers amyF-F and MCS-R and primers amyR-R and MCS-F respectively. Finally, using the Gibson method based on isothermal assembly [67], we joined the three fragments to obtain the PCR product “amy-F–Cm–Promoter–RBS–Luc - amy-R”. The final PCR product was used to transform strain NC57 by selection for cloramphenicol resistance.
Luciferase experiments were performed as we previously described in [68].
All primers are listed in S2 Table.
Construction of the natively expressed gfp-mreB fusion
A method developed to construct scar-less and marker-less deletions in the genome of B. subtilis [69], was adapted to insert the gfp directly upstream mreB, at the native locus.
The first step was to delete, in the recipient strain (NC101, NeoR), the radC gene which is positioned right before mreB, by inserting a deletion cassette (PhleoR). The cassette was first amplified by PCR from plasmid pUC19-K7-010 [69] using the primers K7PH-F and K7PH-R. Then, the regions upstream (radC front) and downstream (radC back) the radC gene were amplified using the primers HindIII-Pmaf-F and Phleo-radC-R or Phleo-radC-F and HindIII-mreB-R, respectively. Finally, the three fragments were joined using the Gibson method [67] to obtain the following PCR product n°1:“radC front–Phleo cassette–radC back”. Transformation of the recipient strain (168 Δupp) with this PCR product generated strain NC102 which is NeoS and PhleoR.
Then, the deletion cassette was replaced by a fragment that re-introduced the radC gene and inserted gfp in front of mreB. This fragment was constituted by two blocks, namely Pmaf-maf-radC (block 1) and gfp-mreB (block 2). These blocks were amplified by PCR using the following primers: Pmaf-F and GFP-radC-R (for block 1) and RBS-mreB-GFP-F and mreB-R (for block 2). The gfp-mreB block was amplified from chromosomal DNA of strain 3723 [41]. The two blocks were then joined using the Gibson method [67] to generate the PCR product n°2: Pmaf-maf-radC-gfp-mreB. This final PCR product was used to transform the strain NC102 to obtain strain NC103 (NeoR and PhleoS), which now contains gfp right in front of mreB inside its own operon. Cells of this strain (NC103) and its derivatives, in which Pnative-gfp-mreB is expressed as the only copy of mreB in the genome, were viable and displayed almost wild-type growth and morphology, indicating that the fusion is virtually functional (S4A Fig and S1 Movie).
All primers used are listed in S2 Table.
Construction of the natively expressed comGA-rfp fusion
We decided to express the comGA-rfp fusion under the control of the native comGA promoter (PcomGA) from the thrC locus. The Gibson method [67] was used to join four PCR fragments corresponding to the upstream (thrC front) and downstream (thrC back) regions of the thrC gene, the comGA promoter and orf, and the mrfpruby gene. These fragments were amplified using the primers hom-F and pDG1664-MCS-R (thrC front), pDG1664-MCS-R and thrB-R (thrC back), pDG1664-MCS-PcomGA-F and RFP-comGA-R (PcomGA-comGA) and comGA-RFP-F and pDG1664-MCS-RFP-R (mrfpruby). The four fragments were joined to produce the final PCR product “thrC front–PcomGA−comGA–mrfpruby - thrC back”. The thrC front and thrC back (which also contains an erythromycin resistance cassette) fragments were amplified from plasmid pDG1664 [70]. The mrfpruby gene was amplified from chromosomal DNA of strain RWSB5 [16]. The final PCR product was used to transform the NC57strain to generate strain NC118. In this strain and its derivatives, ComGA-mRFPruby displays the expected dynamic of localization during competence [4], indicating that the fusion is virtually functional (S5A–S5D Fig).
All primers are listed in S2 Table.
Construction of the Phyperspank-comGA-rfp fusion
The method was comparable to the construction of the natively expressed comGA-rfp fusion described above. The Gibson method [67] was used to join four PCR fragments corresponding to the upstream (thrC front) and downstream (thrC back) regions of the thrC gene, the comGA gene and the mrfpruby gene. The Phyperspank promoter was introduced through the thrC front fragment. The four fragments were amplified using the primers hom-F and pDG1664-MCS-R (thrC front), pDG1664-MCS-R and thrB-R (thrC back), pDG1664-MCS-comGA-F and RFP-comGA-R (comGA) and comGA-RFP-F and pDG1664-MCS-RFP-R (mrfpruby). The four fragments were joined to produce the final PCR product “thrC front–Phyperspank−comGA–mrfpruby - thrC back”. The thrC front (that contains the Phyperspank promoter) and thrC back (that also contains an erythromycin resistance cassette) fragments were amplified from the pDP150 plasmid [71]. The mrfpruby gene was amplified from chromosomal DNA of strain RWSB5 [16]. The final PCR product was used to transform the wild type strain (168) to generating strain NC208.
All primers are listed in S2 Table.
Construction of the PcomK-rfp fusion
We decided to clone the rfp gene under the control of the comK promoter at the amyE locus. The Gibson method [67] was used to join four fragments corresponding to the upstream (amy front) and downstream (amy back) regions of the amyE gene, the comK promoter and the mrfpruby gene. These fragments were amplified using the primers amy-F and PcomK-amy-R (amy front), RFP-amyR-F and amyR-R (amy back), amyF-PcomK-F and RFP-PcomK-R (PcomK) and PcomK-RFP-F and amyR-RFP-R (mrfpruby) respectively. The amy front and back (which also contains a spectinomycin resistance cassette) fragments were amplified from plasmid pDG1730 [70]. The mrfpruby gene was amplified from chromosomal DNA of strain RWSB5 [16]. The four fragments were joined to produce the PCR product “amy front–PcomK−mrfpruby - amy back”. The final PCR product was used to transform the wild type strain (168), inserting the PcomK - mrfpruby construct, at the amyE locus and selecting for chloramphenicol resistance.
All primers are listed in S2 Table.
Construction of the Pxyl-spa-mreB fusion
Translational fusion between the SPA-encoding (Sequential Peptide Affinity) and mreB open reading frame was cloned at the ectopic amyE locus under control of the xylose-inducible promoter Pxyl (pSG-SPA-Nter). pSG-Spa-Nter was generated by replacing the GFP contained in pSG1729 [72] by affinity purification tags (Sequential Peptide Affinity, or SPA) [39], right downstream from the Pxyl promoter. However to generate a N-ter fusion, the tags were inverted in comparison to the original SPA construct (i.e. Flag-TEV site-CBD). The inverted SPA tag was synthesized by Genscript. Then, the mreB open reading frame was PCR-amplified using primers ac-983/ac984, and cloned into the pSG-Spa-Nter vector, using the XhoI and EcoRI restriction sites. The resulting pAC637 plasmid (pSG-Pxyl-spa-mreB) was transformed into B. subtilis strain 4281 (ΔmreB::cm) and selected for resistance to spectinomycin, to obtain strain ABS1370. Finally, we used chromosomal DNA of strain ABS1370 to transfer by natural transformation the amyE::Pxyl-spa-mreB (Spc) construct in strain NC60 to obtain strain NC66.
Pxyl-perR-spa fusion
Chromosomal DNA from strain Bas013 [73] was used to transform the wild-type strain (168) and transfer the Pxyl-perR-spa fusion. Chromosomal DNA of strain NC60 was then used to sequentially incorporate by natural transformation the mcComS and ComK-GFP constructs to generate the final strain NC135.
Luciferase assay
Experiments were carried out as previously described [66,68]. In brief, the high instability of the luciferase, used as transcriptional reporter in B. subtilis, allows us to approach the measurement of a rate of expression [66], with a relatively small contribution from the cumulative effect of transcription. This particular characteristic of luciferase is in stark contrast with the behavior of other reporters, e.g. β-galactosidase. All the strains used in the luciferase experiments carried a multi-copy plasmid, mcComS [74], in order to increase the percentage of competent cells (from 2% to 35% in the wild-type background in the conditions used here, see Fig 4A).
For detection of luciferase activity, strains were first grown in LB medium to an optical density at 600 nm (OD600nm) of 2. Cells were then pelleted and resuspended in fresh competence medium, adjusting all the cultures to an OD600nm of 2. These pre-cultures were then diluted 20 fold in fresh competence medium and 200 µl was distributed in each of two wells in a 96-well black plate (PerkinElmer). 10 µl of luciferin (PerkinElmer) was added to each well to reach a final concentration of 1.5 mg/ml (4.7 mM). The cultures were incubated at 37°C with agitation in a PerkinElmer Envision 2104 Multilabel Reader equipped with an enhanced sensitivity photomultiplier for luminometry. The temperature of the clear plastic lid was maintained at 38°C to avoid condensation. Relative Luminescence Units (RLU) and OD600nm were measured at 2 minutes intervals. The data were plotted as RLU/OD (luminescence readings corrected for the OD) versus time from inoculation.
Transformation efficiency measurements
B. subtilis strains were transformed using chromosomal DNA of strain BD4893 carrying a spectinomycin marker [35]. The number of transformants was evaluated by plating the transformed cultures on LB agar plates containing spectinomycin. Each transformation culture was also plated on non-selective LB agar in dilution series to establish the viable cell count. Transformation efficiency was calculated by dividing the number of transformants by the viable count of each strain.
SPA-tag pull-down experiments
The strains containing the SPA fusions were grown to T2 in competence medium supplemented with 0.4% xylose (to induce the SPA fusions). The cultures were then centrifuged and promptly frozen in liquid nitrogen. The xylose concentration used was chosen in order to optimize the SPA fusions production and minimize the shape and growth phenotypes associated to the over-expression of MreB. The frozen cells pellets were then disrupted by cryogenic grinding (4 cycles of 2 minutes, always maintaining the cupules and the pellets in liquid nitrogen). The powder recovered from the grinding was resuspended in buffer A (Tris-HCl pH7,5 10 mM, NaCl 150 mM, EDTA 0,2 mM, Triton 0,1 mM and proteases inhibitors) and centrifuged to eliminate cell debris. SPA-MreB, PerR-SPA and No-SPA containing protein complexes were then isolated and analyzed as described in [75].
Fluorescence microscopy
Cultures were grown in competence medium at 37°C from single freshly isolated colonies on plates containing the appropriate antibiotic selection. Samples for microscopic observation were taken at T2 (2 hours after the beginning of competence development) and T2+90 (90 minutes after dilution of a T2 culture in fresh competence medium) and immobilized on 1% agarose-coated microscope slides.
Bacteria were imaged with an inverted microscope (Nikon Ti-E) equipped with a 100× oil immersion objective and an environmental chamber maintained at 37°C. Conventional epifluorescence Images were recorded on phase-contrast and fluorescence channels (472/30-nm excitation filter and 520/35-nm emission filter for GFP, 562/40-nm excitation filter and 641/75-nm emission filter for RFP) with an ORCA-R2 camera (Hamamatsu). Images were processed with NIS-Elements (Nikon) software. Exposure time was set up to 200 ms for nativeGFP-MreB and 500 ms for nativeComGA-RFP.
All TIRFM images were acquired on the same inverted microscope with a diode-pumped solid-state laser (Cobolt Calypso, 50mW, 491nm) and an Apo TIRF 100x oil objective (Nikon, NA 1.49). All images were collected with an electron-multiplying charge-coupled device (EMCCD) camera (iXON3 DU-897, Andor) with a gain of 300. Incidence angles and z-position were adjusted individually for all channels to obtain comparable evanescent wave penetration depth and focus position.
Microfluidics
In order to follow B. subtilis growth over time, we used a microfluidic flow chamber technique (CellAsic part of EMD Millipore). The technology is divided in two parts: a perfusion control system and a microfluidic plate (specific for bacteria, B04A) that keeps cells in a single focal plan and allow us to induce and follow events during many generations.
The day before the experiment, strains were grown on selective plates. The next day, cells were resuspended in competence medium to OD = 1. 1µl of this resuspension was used to inoculate 1mL of fresh competence medium. Once the cultures reached early exponential phase, cells were injected in the chamber and incubated under a continuous flow (5µl/hour) of medium at 37°C.
Quantification of the number of ComGA-RFP foci
In order to characterize ComGA-RFP foci at the single cell level, phase contrast and fluorescence images were taken simultaneously for cells grown to stationary phase (T2) in competence media. Fields of view of both images were used to generate sub-images displaying individual cells by applying a two-step algorithm. First, each single cell was detected by applying segmentation to phase-contrast images, resulting sub-images of individual cells with cell contours. Next, diffraction-limited comGA foci in each cell were identified in fluorescence images. Examples of individual cells are presented in S5 Fig Custom image processing codes (S10 Fig) were implemented in Matlab (Mathworks).
GFP-MreB patches speed measurements
Kymograph analysis was applied to obtain the rotation speed of MreB patches as we previously described [16]. In brief, a series of parallel lines were created from one cell pole to the other (every other pixel), all perpendicular to the cell midline. Next, kymographs were generated, corresponding to movement of MreB patches at all positions along the cell longer axis. Finally, angles of the clear MreB traces on the kymographs were used to calculate the rotation speed.
Competent cells length distribution
Length of competent cells was measured using the Metamorph software (Molecular Devices). Phase contrast images were used and the distance from one pole to the other was evaluated. Length of competent cells during the outgrowth experiment is shown as boxplots (refers to Fig 3F). The blue box edges indicate the first and third quartile while the red line indicates the median of the data set. In addition, the whiskers indicate the 5th and 95th percentiles and individual red points indicate outliers. All values with means, standard deviations (SD) and sample sizes are listed in S3 Table. Boxplots were plotted using Matlab 2013. The statistical significance of the differences observed is presented in S3 Table.
Western blot
Whole cell extracts were fractionated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane using a transfer apparatus according to the manufacturer’s protocol (Bio-Rad). After incubation with 5% nonfat milk in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, 0.05% Tween 20) for 60 minutes, membranes were incubated with antibodies against GFP (1 : 10000) overnight at room temperature. Membranes were washed 3 times for 10 minutes with TBST and incubated with a 1 : 10000 dilution of anti-rabbit antibodies for 2h. Blots were washed with TBST three times and developed with the “ECL Prime” kit (Amersham) according to the manufacturer’s protocols. The Chemidoc system (Bio-Rad) was used to reveal the membrane and the Image Lab™ software (Bio-Rad) to analyze the intensity of the bands.
Yeast two-hybrid assay
Saccharomyces cerevisiae cells expressing B. subtilis selected proteins as GAL4 BD fusions were mated with cells expressing either the same or another protein as GAL4 AD fusions as presented in [76]. For each fusion, two independent yeast clones were used. Binary interactions were revealed by growth of diploid cells after 5 days at 30°C on synthetic complete medium lacking leucine, uracil and histidine (to select for expression of the HIS3 interaction reporter, annotated-H). Specific interactions were reproduced independently at least three times.
Supporting Information
Zdroje
1. Dubnau D, Losick R (2006) Bistability in bacteria. Mol Microbiol 61 : 564–572. 16879639
2. Berka RM, Hahn J, Albano M, Draskovic I, Persuh M, et al. (2002) Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol Microbiol 43 : 1331–1345. 11918817
3. Fujita M, Gonzalez-Pastor JE, Losick R (2005) High - and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol 187 : 1357–1368. 15687200
4. Hahn J, Maier B, Haijema BJ, Sheetz M, Dubnau D (2005) Transformation proteins and DNA uptake localize to the cell poles in Bacillus subtilis. Cell 122 : 59–71. 16009133
5. Higgins D, Dworkin J (2012) Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 36 : 131–148. doi: 10.1111/j.1574-6976.2011.00310.x 22091839
6. Ogura M, Yamaguchi H, Kobayashi K, Ogasawara N, Fujita Y, et al. (2002) Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J Bacteriol 184 : 2344–2351. 11948146
7. Hamoen LW, Smits WK, de Jong A, Holsappel S, Kuipers OP (2002) Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res 30 : 5517–5528. 12490720
8. Cahn FH, Fox MS (1968) Fractionation of transformable bacteria from ocompetent cultures of Bacillus subtilis on renografin gradients. J Bacteriol 95 : 867–875. 4966830
9. Hadden C, Nester EW (1968) Purification of competent cells in the Bacillus subtilis transformation system. J Bacteriol 95 : 876–885. 4966831
10. Hahn J, Albano M, Dubnau D (1987) Isolation and characterization of Tn917lac-generated competence mutants of Bacillus subtilis. J Bacteriol 169 : 3104–3109. 3036770
11. Haijema BJ, Hahn J, Haynes J, Dubnau D (2001) A ComGA-dependent checkpoint limits growth during the escape from competence. Mol Microbiol 40 : 52–64. 11298275
12. Briley K Jr., Prepiak P, Dias MJ, Hahn J, Dubnau D (2011) Maf acts downstream of ComGA to arrest cell division in competent cells of B. subtilis. Mol Microbiol 81 : 23–39. doi: 10.1111/j.1365-2958.2011.07695.x 21564336
13. Kramer N, Hahn J, Dubnau D (2007) Multiple interactions among the competence proteins of Bacillus subtilis. Mol Microbiol 65 : 454–464. 17630974
14. Chastanet A, Carballido-López R (2012) The actin-like MreB proteins in Bacillus subtilis: a new turn. Front Biosci (Schol Ed) 4 : 1582–1606. 22652894
15. Carballido-López R (2006) The bacterial actin-like cytoskeleton. Microbiol Mol Biol Rev 70 : 888–909. 17158703
16. Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, et al. (2011) Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333 : 225–228. doi: 10.1126/science.1203466 21636744
17. Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, et al. (2011) Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333 : 222–225. doi: 10.1126/science.1203285 21636745
18. Olshausen PV, Defeu Soufo HJ, Wicker K, Heintzmann R, Graumann PL, et al. (2013) Superresolution Imaging of Dynamic MreB Filaments in B. subtilis-A Multiple-Motor-Driven Transport? Biophys J 105 : 1171–1181. doi: 10.1016/j.bpj.2013.07.038 24010660
19. Reimold C, Defeu Soufo HJ, Dempwolff F, Graumann PL (2013) Motion of variable-length MreB filaments at the bacterial cell membrane influences cell morphology. Mol Biol Cell 24 : 2340–2349. doi: 10.1091/mbc.E12-10-0728 23783036
20. Defeu Soufo HJ, Graumann PL (2004) Dynamic movement of actin-like proteins within bacterial cells. EMBO Rep 5 : 789–794. 15272301
21. van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, et al. (2011) The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci U S A 108 : 15822–15827. doi: 10.1073/pnas.1108999108 21903929
22. Kawai Y, Asai K, Errington J (2009) Partial functional redundancy of MreB isoforms, MreB, Mbl and MreBH, in cell morphogenesis of Bacillus subtilis. Mol Microbiol 73 : 719–731. doi: 10.1111/j.1365-2958.2009.06805.x 19659933
23. Jones LJ, Carballido-Lopez R, Errington J (2001) Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104 : 913–922. 11290328
24. Schirner K, Errington J (2009) Influence of heterologous MreB proteins on cell morphology of Bacillus subtilis. Microbiology 155 : 3611–3621. doi: 10.1099/mic.0.030692-0 19643765
25. Carballido-López R, Formstone A, Li Y, Ehrlich SD, Noirot P, et al. (2006) Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell 11 : 399–409. 16950129
26. Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, et al. (2012) Condition-Dependent Transcriptome Reveals High-Level Regulatory Architecture in Bacillus subtilis. Science 335 : 1103–1106. doi: 10.1126/science.1206848 22383849
27. Eiamphungporn W, Helmann JD (2008) The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses. Mol Microbiol 67 : 830–848. doi: 10.1111/j.1365-2958.2007.06090.x 18179421
28. Kunkel B, Kroos L, Poth H, Youngman P, Losick R (1989) Temporal and spatial control of the mother-cell regulatory gene spoIIID of Bacillus subtilis. Genes Dev 3 : 1735–1744. 2514119
29. Decatur A, McMurry MT, Kunkel BN, Losick R (1997) Translation of the mRNA for the sporulation gene spoIIID of Bacillus subtilis is dependent upon translation of a small upstream open reading frame. J Bacteriol 179 : 1324–1328. 9023218
30. Feucht A, Evans L, Errington J (2003) Identification of sporulation genes by genome-wide analysis of the sigmaE regulon of Bacillus subtilis. Microbiology 149 : 3023–3034. 14523133
31. Tseng CL, Shaw GC (2008) Genetic evidence for the actin homolog gene mreBH and the bacitracin resistance gene bcrC as targets of the alternative sigma factor SigI of Bacillus subtilis. J Bacteriol 190 : 1561–1567. 18156261
32. Turgay K, Hahn J, Burghoorn J, Dubnau D (1998) Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J 17 : 6730–6738. 9890793
33. Kong L, Siranosian KJ, Grossman AD, Dubnau D (1993) Sequence and properties of mecA, a negative regulator of genetic competence in Bacillus subtilis. Mol Microbiol 9 : 365–373. 8412687
34. Asai K, Yamaguchi H, Kang CM, Yoshida K, Fujita Y, et al. (2003) DNA microarray analysis of Bacillus subtilis sigma factors of extracytoplasmic function family. FEMS Microbiol Lett 220 : 155–160. 12644242
35. Albano M, Hahn J, Dubnau D (1987) Expression of competence genes in Bacillus subtilis. J Bacteriol 169 : 3110–3117. 3110135
36. Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, et al. (2004) The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol 2: e328. 15383836
37. Hahn J, Bylund J, Haines M, Higgins M, Dubnau D (1995) Inactivation of mecA prevents recovery from the competent state and interferes with cell division and the partitioning of nucleoids in Bacillus subtilis. Mol Microbiol 18 : 755–767. 8817496
38. Rashid MH, Tamakoshi A, Sekiguchi J (1996) Effects of mecA and mecB (clpC) mutations on expression of sigD, which encodes an alternative sigma factor, and autolysin operons and on flagellin synthesis in Bacillus subtilis. J Bacteriol 178 : 4861–4869. 8759849
39. Zeghouf M, Li J, Butland G, Borkowska A, Canadien V, et al. (2004) Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J Proteome Res 3 : 463–468. 15253427
40. Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, et al. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4 : 1265–1272. 15958392
41. Formstone A, Errington J (2005) A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol Microbiol 55 : 1646–1657. 15752190
42. Schirner K, Errington J (2009) The cell wall regulator {sigma}I specifically suppresses the lethal phenotype of mbl mutants in Bacillus subtilis. J Bacteriol 191 : 1404–1413. doi: 10.1128/JB.01497-08 19114499
43. Bruckner A, Polge C, Lentze N, Auerbach D, Schlattner U (2009) Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci 10 : 2763–2788. doi: 10.3390/ijms10062763 19582228
44. Kawai Y, Daniel RA, Errington J (2009) Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol 71 : 1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x 19192185
45. Dubnau D (1997) Binding and transport of transforming DNA by Bacillus subtilis: the role of type-IV pilin-like proteins—a review. Gene 192 : 191–198. 9224890
46. Jakutyte L, Baptista C, Sao-Jose C, Daugelavicius R, Carballido-Lopez R, et al. (2011) Bacteriophage infection in rod-shaped gram-positive bacteria: evidence for a preferential polar route for phage SPP1 entry in Bacillus subtilis. J Bacteriol 193 : 4893–4903. doi: 10.1128/JB.05104-11 21705600
47. Sao-Jose C, Baptista C, Santos MA (2004) Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J Bacteriol 186 : 8337–8346. 15576783
48. Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, et al. (2007) Type VII secretion—mycobacteria show the way. Nat Rev Microbiol 5 : 883–891. 17922044
49. Nilsen T, Yan AW, Gale G, Goldberg MB (2005) Presence of multiple sites containing polar material in spherical Escherichia coli cells that lack MreB. J Bacteriol 187 : 6187–6196. 16109960
50. Pradel N, Santini CL, Bernadac A, Shih YL, Goldberg MB, et al. (2007) Polar positional information in Escherichia coli spherical cells. Biochem Biophys Res Commun 353 : 493–500. 17188233
51. Cowles KN, Gitai Z (2010) Surface association and the MreB cytoskeleton regulate pilus production, localization and function in Pseudomonas aeruginosa. Mol Microbiol 76 : 1411–1426. doi: 10.1111/j.1365-2958.2010.07132.x 20398206
52. Briley K Jr., Dorsey-Oresto A, Prepiak P, Dias MJ, Mann JM, et al. (2011) The secretion ATPase ComGA is required for the binding and transport of transforming DNA. Mol Microbiol 81 : 818–830. doi: 10.1111/j.1365-2958.2011.07730.x 21707789
53. Rudner DZ, Losick R (2010) Protein subcellular localization in bacteria. Cold Spring Harb Perspect Biol 2: a000307. doi: 10.1101/cshperspect.a000307 20452938
54. Siripala AD, Welch MD (2007) SnapShot: actin regulators II. Cell 128 : 1014. 17350583
55. Siripala AD, Welch MD (2007) SnapShot: actin regulators I. Cell 128 : 626. 17289579
56. Schirner K, Eun YJ, Dion M, Luo Y, Helmann JD, et al. (2015) Lipid-linked cell wall precursors regulate membrane association of bacterial actin MreB. Nat Chem Biol 11 : 38–45. doi: 10.1038/nchembio.1689 25402772
57. Lam H, Oh DC, Cava F, Takacs CN, Clardy J, et al. (2009) D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325 : 1552–1555. doi: 10.1126/science.1178123 19762646
58. Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J (1999) The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci U S A 96 : 14819–14824. 10611296
59. Gueiros-Filho FJ, Losick R (2002) A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev 16 : 2544–2556. 12368265
60. Romberg L, Levin PA (2003) Assembly dynamics of the bacterial cell division protein FTSZ: poised at the edge of stability. Annu Rev Microbiol 57 : 125–154. 14527275
61. Wu LJ, Errington J (2004) Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117 : 915–925. 15210112
62. Haeusser DP, Schwartz RL, Smith AM, Oates ME, Levin PA (2004) EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol Microbiol 52 : 801–814. 15101985
63. Bendezu FO, Hale CA, Bernhardt TG, de Boer PA (2009) RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J 28 : 193–204. doi: 10.1038/emboj.2008.264 19078962
64. Huang WZ, Wang JJ, Chen HJ, Chen JT, Shaw GC (2013) The heat-inducible essential response regulator WalR positively regulates transcription of sigI, mreBH and lytE in Bacillus subtilis under heat stress. Res Microbiol 164 : 998–1008. doi: 10.1016/j.resmic.2013.10.003 24125693
65. Saito H, Miura KI (1963) Preparation of Transforming Deoxyribonucleic Acid by Phenol Treatment. Biochim Biophys Acta 72 : 619–629. 14071565
66. Mirouze N, Prepiak P, Dubnau D (2011) Fluctuations in spo0A transcription control rare developmental transitions in Bacillus subtilis. PLoS Genet 7: e1002048. doi: 10.1371/journal.pgen.1002048 21552330
67. Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, et al. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6 : 343–345. doi: 10.1038/nmeth.1318 19363495
68. Mirouze N, Desai Y, Raj A, Dubnau D (2012) Spo0A~P imposes a temporal gate for the bimodal expression of competence in Bacillus subtilis. PLoS Genet 8: e1002586. doi: 10.1371/journal.pgen.1002586 22412392
69. Fabret C, Ehrlich SD, Noirot P (2002) A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol Microbiol 46 : 25–36. 12366828
70. Guerout-Fleury AM, Frandsen N, Stragier P (1996) Plasmids for ectopic integration in Bacillus subtilis. Gene 180 : 57–61. 8973347
71. Kearns DB, Losick R (2005) Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev 19 : 3083–3094. 16357223
72. Lewis PJ, Marston AL (1999) GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227 : 101–110. 9931458
73. Rueff AS, Chastanet A, Dominguez-Escobar J, Yao Z, Yates J, et al. (2014) An early cytoplasmic step of peptidoglycan synthesis is associated to MreB in Bacillus subtilis. Mol Microbiol 91 : 348–362. doi: 10.1111/mmi.12467 24261876
74. Hahn J, Luttinger A, Dubnau D (1996) Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis. Mol Microbiol 21 : 763–775. 8878039
75. Delumeau O, Lecointe F, Muntel J, Guillot A, Guedon E, et al. (2011) The dynamic protein partnership of RNA polymerase in Bacillus subtilis. Proteomics 11 : 2992–3001. doi: 10.1002/pmic.201000790 21710567
76. Marchadier E, Carballido-Lopez R, Brinster S, Fabret C, Mervelet P, et al. (2011) An expanded protein-protein interaction network in Bacillus subtilis reveals a group of hubs: Exploration by an integrative approach. Proteomics 11 : 2981–2991. doi: 10.1002/pmic.201000791 21630458
Štítky
Genetika Reprodukční medicína
Článek Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and ThrombocytopeniaČlánek Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of andČlánek Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on PhenotypeČlánek Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human CancersČlánek A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic BacteriaČlánek The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System inČlánek The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological LifespanČlánek The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 6
-
Všechny články tohoto čísla
- Expression of Concern: RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Orphan Genes Find a Home: Interspecific Competition and Gene Network Evolution
- Minor Cause—Major Effect: A Novel Mode of Control of Bistable Gene Expression
- Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and Thrombocytopenia
- Leveraging Identity-by-Descent for Accurate Genotype Inference in Family Sequencing Data
- Is Required for the Expression of Principal Recognition Molecules That Control Axon Targeting in the Retina
- Epigenetic Aging Signatures Are Coherently Modified in Cancer
- Silencing of DNase Colicin E8 Gene Expression by a Complex Nucleoprotein Assembly Ensures Timely Colicin Induction
- A Transposable Element within the Non-canonical Telomerase RNA of Modulates Telomerase in Response to DNA Damage
- The Orphan Gene Regulates Dauer Development and Intraspecific Competition in Nematodes by Copy Number Variation
- 9--13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice
- The DnaA Protein Is Not the Limiting Factor for Initiation of Replication in
- FGFR3 Deficiency Causes Multiple Chondroma-like Lesions by Upregulating Hedgehog Signaling
- Multiple Changes of Gene Expression and Function Reveal Genomic and Phenotypic Complexity in SLE-like Disease
- Directed Evolution of RecA Variants with Enhanced Capacity for Conjugational Recombination
- The Regulatory T Cell Lineage Factor Foxp3 Regulates Gene Expression through Several Distinct Mechanisms Mostly Independent of Direct DNA Binding
- MreB-Dependent Inhibition of Cell Elongation during the Escape from Competence in
- DNA Damage Regulates Translation through β-TRCP Targeting of CReP
- Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of and
- The Analysis of () Mutants Reveals Differences in the Fusigenic Potential among Telomeres
- The Causative Gene in Chanarian Dorfman Syndrome Regulates Lipid Droplet Homeostasis in .
- Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on Phenotype
- The . Accessory Helicase PcrA Facilitates DNA Replication through Transcription Units
- AKTIP/Ft1, a New Shelterin-Interacting Factor Required for Telomere Maintenance
- Npvf: Hypothalamic Biomarker of Ambient Temperature Independent of Nutritional Status
- Transfer RNAs Mediate the Rapid Adaptation of to Oxidative Stress
- Connecting Circadian Genes to Neurodegenerative Pathways in Fruit Flies
- Response to “Ribosome Rescue and Translation Termination at Non-standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Response to the Formal Letter of Z. Chrzanowska-Lightowlers and R. N. Lightowlers Regarding Our Article “Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human Cancers
- Regulation of Insulin Receptor Trafficking by Bardet Biedl Syndrome Proteins
- Altered Levels of Mitochondrial DNA Are Associated with Female Age, Aneuploidy, and Provide an Independent Measure of Embryonic Implantation Potential
- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Canine Spontaneous Head and Neck Squamous Cell Carcinomas Represent Their Human Counterparts at the Molecular Level
- Genetic Changes to a Transcriptional Silencer Element Confers Phenotypic Diversity within and between Species
- Functional Assessment of Disease-Associated Regulatory Variants Using a Versatile Dual Colour Transgenesis Strategy in Zebrafish
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Independent Neuronal Origin of Seizures and Behavioral Comorbidities in an Animal Model of a Severe Childhood Genetic Epileptic Encephalopathy
- The Human Blood Metabolome-Transcriptome Interface
- A Common Cancer Risk-Associated Allele in the Locus Encodes a Dominant Negative Inhibitor of Telomerase
- A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic Bacteria
- The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System in
- The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological Lifespan
- Dissection of a Complex Disease Susceptibility Region Using a Bayesian Stochastic Search Approach to Fine Mapping
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
- Ancestral Chromatin Configuration Constrains Chromatin Evolution on Differentiating Sex Chromosomes in
- Abnormal Activation of BMP Signaling Causes Myopathy in Null Mice
- Reproductive Mode and the Evolution of Genome Size and Structure in Nematodes
- Replication and Active Partition of Integrative and Conjugative Elements (ICEs) of the SXT/R391 Family: The Line between ICEs and Conjugative Plasmids Is Getting Thinner
- Motor and Sensory Deficits in the Mice Result from Mutation of the ESCRT Component HGS
- Senescence in the Sbds-Deficient Murine Pancreas: Cell-Type Specific Consequences of Translation Insufficiency
- Lipophorin Receptors Recruit the Lipoprotein LTP to the Plasma Membrane to Mediate Lipid Uptake
- Separable Crossover-Promoting and Crossover-Constraining Aspects of Zip1 Activity during Budding Yeast Meiosis
- Context-Dependent Functional Divergence of the Notch Ligands DLL1 and DLL4
- Multilayered Organization of Jasmonate Signalling in the Regulation of Root Growth
- Necrotic Cells Actively Attract Phagocytes through the Collaborative Action of Two Distinct PS-Exposure Mechanisms
- A Novel Feedback Loop That Controls Bimodal Expression of Genetic Competence
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Human Blood Metabolome-Transcriptome Interface
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



