-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaOrphan Genes Find a Home: Interspecific Competition and Gene Network Evolution
article has not abstract
Published in the journal: . PLoS Genet 11(6): e32767. doi:10.1371/journal.pgen.1005254
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1005254Summary
article has not abstract
Conservation of gene and protein sequence, and, therefore, conservation of the resulting molecular interactions that mediate biological processes, is foundational to our understanding of biology. This conservation allows discovery in one organism, such as worms or mice, to inform our understanding of the biology in another organism, such as humans. However, there is emerging recognition that many biological processes involve important non-conserved elements, and that de novo gene birth provides an important mechanism for functional evolution [1–3]. Understanding how these novel elements incorporate into gene regulatory networks and alter the network architecture is an important area for theoretical research [4–5], but few experimental examples have been described.
Formation of the dauer larva by nematodes is an adaptation that is responsive to a variety of environmental cues and alterations, features that suggest the underlying gene regulatory network might benefit from enhanced robustness and evolvability [6]. The dauer larva is a relatively dormant, alternative developmental stage that nematodes enter under stressful conditions, such as low food or crowding (signaled by pheromone), that confers increased longevity and tolerance to stressors [7]. In many parasitic nematodes, this stage corresponds to the infective larval stage when the larvae transition to new hosts. There are important conserved components of the gene network that regulate dauer formation, one being the nuclear hormone receptor DAF-12, which is important in free-living as well as parasitic nematode species [8–10]. However, it is also clear that this pathway is subject to considerable inter - and intraspecific differences [11–13]. Recent work on natural isolates of the nematode Pristionchus pacificus identified strain-specific phenotypic differences in dauer formation, and argued that genetically distinct populations exhibit greater sensitivity to pheromone from other populations (pheromone cross-preference [14]). The genetic alterations responsible for these phenotypic differences, however, had not yet been identified.
A new paper in this issue of PLOS Genetics [15] provides an answer, and at the same time highlights how “orphan” genes can be incorporated into conserved regulatory networks. The authors started with two strains of P. pacificus that exhibited marked differences in response to pheromone signaling: the RS2333/California strain, with low dauer formation in response to pheromone, and the RS5134/Ohio strain, with a high response. They then generated 911 recombinant inbred lines (RIL) between the strains, and used Quantitative-Trait-Loci (QTL) mapping to identify a novel gene with no apparent orthologs outside of Pristionchus, dauerless (dau-1), that had undergone a duplication event in the RS2333/California strain. This suggested that dau-1 functions as a repressor of dauer formation in a dose-dependent manner. To test this hypothesis, the authors created transgenic lines with multiple copies of dau-1, and created deletion mutants using CRISPR/Cas9 technology. Their data support the hypothesis as P. pacificus animals with more than two gene copies show no dauer formation in response to pheromone, whereas the deletion of one or both gene copies in RS2333 results in significantly higher dauer formation. The authors also show that this function is mediated by the CAN neurons. dau-1 is expressed in these cells, and ablation of these neurons results in increased dauer formation similar to that seen in dau-1 mutants. Finally, epistasis experiments show that dau-1 acts either downstream or in parallel to steroid hormone signaling, but is dependent on the nuclear hormone receptor gene daf-12 (Fig 1).
Fig. 1. Proposed model for genetic evolution in P. pacificus dauer formation. 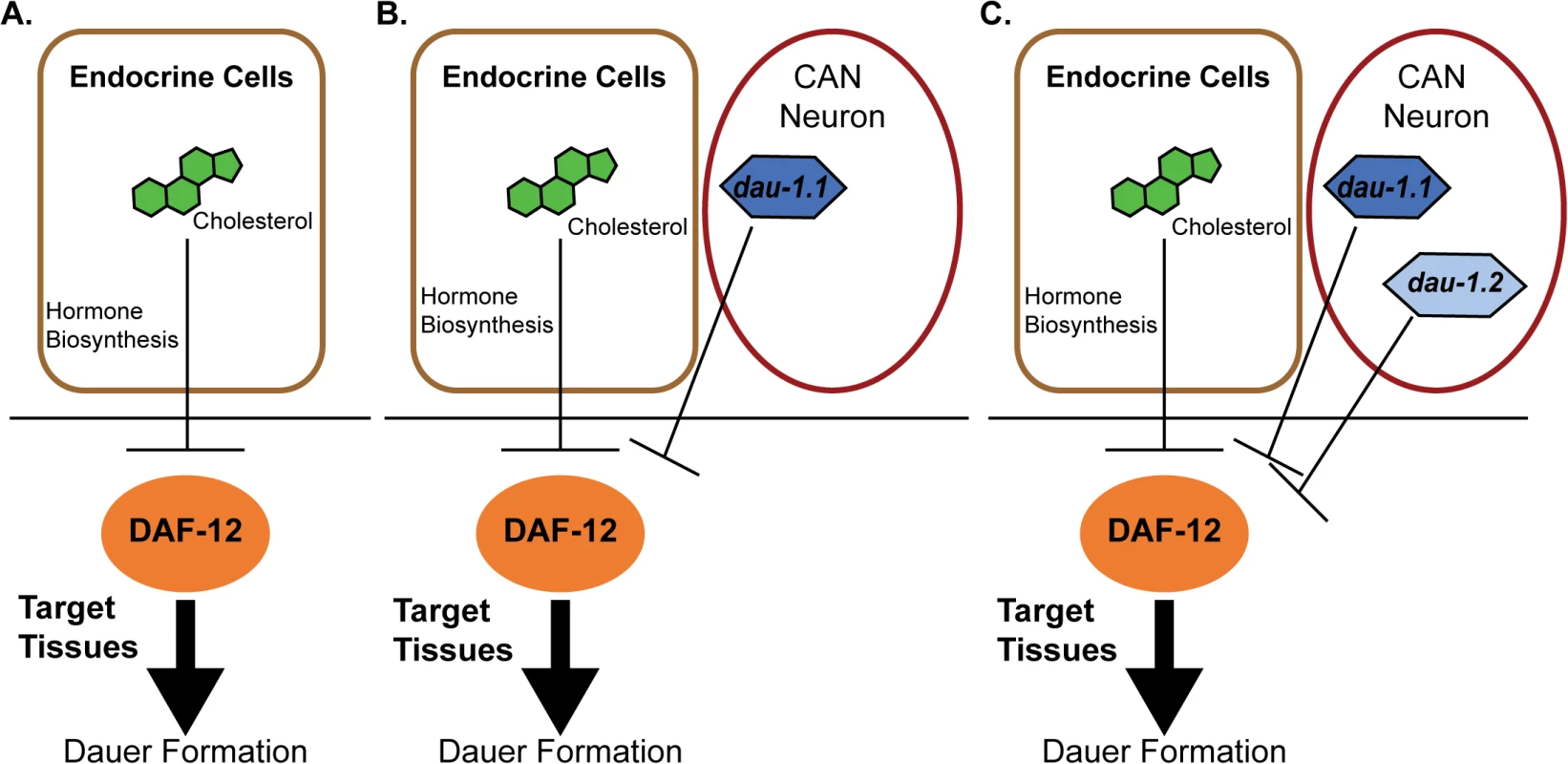
A) Conserved features of the dauer formation pathway. In endocrine cells, cholesterol is a substrate for the biosynthesis of dafachronic acid (DA), a DAF-12 ligand that suppresses the ability of DAF-12 to promote dauer formation. B) Model for the RS5134/Ohio strain with one copy of dauerless (dau-1.1). dau-1.1 is expressed in the CAN neuron cells and acts (genetically) to inhibit DAF-12 function and thereby inhibit dauer formation. C) The RS2333/California strain has two copies of dauerless (dau-1.1 and dau-1.2), and a corresponding double effect of inhibition of dauer formation. Discovery of the dau-1 genes provides insight for how orphan genes can play an important role in the function and the evolution of biological networks that are conserved across species. The presence—and the duplication—of this gene is interpreted to provide a selective advantage in the context of intraspecific competition because it would allow individuals to continue with a reproductive life cycle even in the presence of crowding. Indeed, Mayer et al. have uncovered additional dau-1 paralogs in RS2333, indicating a potential for ongoing duplication of these novel genes [15]. A second important finding of this paper is that these orphan genes influence dauer formation by modulating a pathway with highly conserved elements (Fig 1). The results provide an important experimental example to complement the theoretical models for how the evolution of novel genes can add functional modifications to conserved regulatory networks. In this case, orphan gene evolution contributes to the evolutionary arms race between competing strains.
Important questions remain. In particular, previous work showed that different P. pacificus isolates exhibit differences in pheromone signaling and dauer survival, in addition to pheromone response, arguing that there are additional genetic modifications in the dauer regulatory network [14]. Whether these functions are influenced by dau-1 or related genes, and how these features of the dauer regulation network may interact, is not known. In addition, whether this represents a unique example or an evolutionary prototype is not clear. Generalizations of the types of changes that are responsible for evolutionary change have focused on the level of individual orthologous (and, therefore, conserved) genes [16–17]. These earlier analyses did, however, highlight how the network position of a gene can influence whether it is likely subject to evolutionary modification, and the types of network nodes that are more likely to be affected by cis-regulatory or protein-coding changes. As more evolutionary examples involving orphan genes are described, it will be interesting to learn if they are preferentially incorporated into particular nodes of regulatory networks, and whether they contribute primarily to intraspecific differences or have a role at longer evolutionary timescales.
Zdroje
1. Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12 : 692–702. doi: 10.1038/nrg3053 21878963
2. Carvunis A-R, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, et al. Proto-genes and de novo gene birth. Nature. 2012;487 : 370–374. doi: 10.1038/nature11184 22722833
3. Abrusán G. Integration of new genes into cellular networks, and their structural maturation. Genetics. 2013;195 : 1407–1417. doi: 10.1534/genetics.113.152256 24056411
4. Aldana M, Balleza E, Kauffman S, Resendiz O. Robustness and evolvability in genetic regulatory networks. J Theor Biol. 2007;245 : 433–448. 17188715
5. Pechenick DA, Moore JH, Payne JL. The influence of assortativity on the robustness and evolvability of gene regulatory networks upon gene birth. J Theor Biol. 2013;330 : 26–36. doi: 10.1016/j.jtbi.2013.03.019 23542384
6. Crombach A, Hogeweg P. Evolution of evolvability in gene regulatory networks. PLOS Comput Biol. 2008;4: e1000112. doi: 10.1371/journal.pcbi.1000112 18617989
7. Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22 : 2149–2165. doi: 10.1101/gad.1701508 18708575
8. Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14 : 1512–1527. 10859169
9. Ogawa A, Streit A, Antebi A, Sommer RJ. A conserved endocrine mechanism controls the formation of dauer and infective larvae in nematodes. Curr Biol CB. 2009;19 : 67–71. doi: 10.1016/j.cub.2008.11.063 19110431
10. Wang Z, Zhou XE, Motola DL, Gao X, Suino-Powell K, Conneely A, et al. Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes. Proc Natl Acad Sci U S A. 2009;106 : 9138–9143. doi: 10.1073/pnas.0904064106 19497877
11. Elling AA, Mitreva M, Recknor J, Gai X, Martin J, Maier TR, et al. Divergent evolution of arrested development in the dauer stage of Caenorhabditis elegans and the infective stage of Heterodera glycines. Genome Biol. 2007;8: R211. 17919324
12. Stoltzfus JD, Minot S, Berriman M, Nolan TJ, Lok JB. RNAseq analysis of the parasitic nematode Strongyloides stercoralis reveals divergent regulation of canonical dauer pathways. PLOS Negl Trop Dis. 2012;6: e1854. doi: 10.1371/journal.pntd.0001854 23145190
13. Crook M. The dauer hypothesis and the evolution of parasitism: 20 years on and still going strong. Int J Parasitol. 2014;44 : 1–8. doi: 10.1016/j.ijpara.2013.08.004 24095839
14. Bose N, Meyer JM, Yim JJ, Mayer MG, Markov GV, Ogawa A, et al. Natural variation in dauer pheromone production and sensing supports intraspecific competition in nematodes. Curr Biol CB. 2014;24 : 1536–1541. doi: 10.1016/j.cub.2014.05.045 24980503
15. Mayer MG, Rodelsperger C, Witte H, Riebesell M, Sommer RJ. The Orphan Gene dauerless Regulates Dauer Development and Intraspecific Competition in Nematodes by Copy Number Variation. PLoS Genet 11(6): e1005146. doi: 10.1371/journal.pgen.1005146
16. Stern DL, Orgogozo V. The loci of evolution: how predictable is genetic evolution? Evol Int J Org Evol. 2008;62 : 2155–2177.
17. Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323 : 746–751. doi: 10.1126/science.1158997 19197055
Štítky
Genetika Reprodukční medicína
Článek Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and ThrombocytopeniaČlánek Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of andČlánek Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on PhenotypeČlánek Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human CancersČlánek A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic BacteriaČlánek The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System inČlánek The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological LifespanČlánek The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 6
-
Všechny články tohoto čísla
- Expression of Concern: RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Orphan Genes Find a Home: Interspecific Competition and Gene Network Evolution
- Minor Cause—Major Effect: A Novel Mode of Control of Bistable Gene Expression
- Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and Thrombocytopenia
- Leveraging Identity-by-Descent for Accurate Genotype Inference in Family Sequencing Data
- Is Required for the Expression of Principal Recognition Molecules That Control Axon Targeting in the Retina
- Epigenetic Aging Signatures Are Coherently Modified in Cancer
- Silencing of DNase Colicin E8 Gene Expression by a Complex Nucleoprotein Assembly Ensures Timely Colicin Induction
- A Transposable Element within the Non-canonical Telomerase RNA of Modulates Telomerase in Response to DNA Damage
- The Orphan Gene Regulates Dauer Development and Intraspecific Competition in Nematodes by Copy Number Variation
- 9--13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice
- The DnaA Protein Is Not the Limiting Factor for Initiation of Replication in
- FGFR3 Deficiency Causes Multiple Chondroma-like Lesions by Upregulating Hedgehog Signaling
- Multiple Changes of Gene Expression and Function Reveal Genomic and Phenotypic Complexity in SLE-like Disease
- Directed Evolution of RecA Variants with Enhanced Capacity for Conjugational Recombination
- The Regulatory T Cell Lineage Factor Foxp3 Regulates Gene Expression through Several Distinct Mechanisms Mostly Independent of Direct DNA Binding
- MreB-Dependent Inhibition of Cell Elongation during the Escape from Competence in
- DNA Damage Regulates Translation through β-TRCP Targeting of CReP
- Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of and
- The Analysis of () Mutants Reveals Differences in the Fusigenic Potential among Telomeres
- The Causative Gene in Chanarian Dorfman Syndrome Regulates Lipid Droplet Homeostasis in .
- Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on Phenotype
- The . Accessory Helicase PcrA Facilitates DNA Replication through Transcription Units
- AKTIP/Ft1, a New Shelterin-Interacting Factor Required for Telomere Maintenance
- Npvf: Hypothalamic Biomarker of Ambient Temperature Independent of Nutritional Status
- Transfer RNAs Mediate the Rapid Adaptation of to Oxidative Stress
- Connecting Circadian Genes to Neurodegenerative Pathways in Fruit Flies
- Response to “Ribosome Rescue and Translation Termination at Non-standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Response to the Formal Letter of Z. Chrzanowska-Lightowlers and R. N. Lightowlers Regarding Our Article “Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human Cancers
- Regulation of Insulin Receptor Trafficking by Bardet Biedl Syndrome Proteins
- Altered Levels of Mitochondrial DNA Are Associated with Female Age, Aneuploidy, and Provide an Independent Measure of Embryonic Implantation Potential
- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Canine Spontaneous Head and Neck Squamous Cell Carcinomas Represent Their Human Counterparts at the Molecular Level
- Genetic Changes to a Transcriptional Silencer Element Confers Phenotypic Diversity within and between Species
- Functional Assessment of Disease-Associated Regulatory Variants Using a Versatile Dual Colour Transgenesis Strategy in Zebrafish
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Independent Neuronal Origin of Seizures and Behavioral Comorbidities in an Animal Model of a Severe Childhood Genetic Epileptic Encephalopathy
- The Human Blood Metabolome-Transcriptome Interface
- A Common Cancer Risk-Associated Allele in the Locus Encodes a Dominant Negative Inhibitor of Telomerase
- A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic Bacteria
- The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System in
- The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological Lifespan
- Dissection of a Complex Disease Susceptibility Region Using a Bayesian Stochastic Search Approach to Fine Mapping
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
- Ancestral Chromatin Configuration Constrains Chromatin Evolution on Differentiating Sex Chromosomes in
- Abnormal Activation of BMP Signaling Causes Myopathy in Null Mice
- Reproductive Mode and the Evolution of Genome Size and Structure in Nematodes
- Replication and Active Partition of Integrative and Conjugative Elements (ICEs) of the SXT/R391 Family: The Line between ICEs and Conjugative Plasmids Is Getting Thinner
- Motor and Sensory Deficits in the Mice Result from Mutation of the ESCRT Component HGS
- Senescence in the Sbds-Deficient Murine Pancreas: Cell-Type Specific Consequences of Translation Insufficiency
- Lipophorin Receptors Recruit the Lipoprotein LTP to the Plasma Membrane to Mediate Lipid Uptake
- Separable Crossover-Promoting and Crossover-Constraining Aspects of Zip1 Activity during Budding Yeast Meiosis
- Context-Dependent Functional Divergence of the Notch Ligands DLL1 and DLL4
- Multilayered Organization of Jasmonate Signalling in the Regulation of Root Growth
- Necrotic Cells Actively Attract Phagocytes through the Collaborative Action of Two Distinct PS-Exposure Mechanisms
- A Novel Feedback Loop That Controls Bimodal Expression of Genetic Competence
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Human Blood Metabolome-Transcriptome Interface
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



