-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaStructures of Receptor Complexes of a North American H7N2 Influenza Hemagglutinin with a Loop Deletion in the Receptor Binding Site
Human infections with subtype H7 avian influenza viruses have been reported as early as 1979. In 1996, a genetically stable 24-nucleotide deletion emerged in North American H7 influenza virus hemagglutinins, resulting in an eight amino acid deletion in the receptor-binding site. The continuous circulation of these viruses in live bird markets, as well as its documented ability to infect humans, raises the question of how these viruses achieve structural stability and functionality. Here we report a detailed molecular analysis of the receptor binding site of the North American lineage subtype H7N2 virus A/New York/107/2003 (NY107), including complexes with an avian receptor analog (3′-sialyl-N-acetyllactosamine, 3′SLN) and two human receptor analogs (6′-sialyl-N-acetyllactosamine, 6′SLN; sialyllacto-N-tetraose b, LSTb). Structural results suggest a novel mechanism by which residues Arg220 and Arg229 (H3 numbering) are used to compensate for the deletion of the 220-loop and form interactions with the receptor analogs. Glycan microarray results reveal that NY107 maintains an avian-type (α2-3) receptor binding profile, with only moderate binding to human-type (α2-6) receptor. Thus despite its dramatically altered receptor binding site, this HA maintains functionality and confirms a need for continued influenza virus surveillance of avian and other animal reservoirs to define their zoonotic potential.
Published in the journal: . PLoS Pathog 6(9): e32767. doi:10.1371/journal.ppat.1001081
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001081Summary
Human infections with subtype H7 avian influenza viruses have been reported as early as 1979. In 1996, a genetically stable 24-nucleotide deletion emerged in North American H7 influenza virus hemagglutinins, resulting in an eight amino acid deletion in the receptor-binding site. The continuous circulation of these viruses in live bird markets, as well as its documented ability to infect humans, raises the question of how these viruses achieve structural stability and functionality. Here we report a detailed molecular analysis of the receptor binding site of the North American lineage subtype H7N2 virus A/New York/107/2003 (NY107), including complexes with an avian receptor analog (3′-sialyl-N-acetyllactosamine, 3′SLN) and two human receptor analogs (6′-sialyl-N-acetyllactosamine, 6′SLN; sialyllacto-N-tetraose b, LSTb). Structural results suggest a novel mechanism by which residues Arg220 and Arg229 (H3 numbering) are used to compensate for the deletion of the 220-loop and form interactions with the receptor analogs. Glycan microarray results reveal that NY107 maintains an avian-type (α2-3) receptor binding profile, with only moderate binding to human-type (α2-6) receptor. Thus despite its dramatically altered receptor binding site, this HA maintains functionality and confirms a need for continued influenza virus surveillance of avian and other animal reservoirs to define their zoonotic potential.
Introduction
Influenza is an acute respiratory virus that infects up to 20% of the population in the United States, resulting in ∼36,000 deaths annually [1], [2]. The two membrane glycoproteins on the surface of influenza A virus, hemagglutinin (HA), which functions as the receptor binding and membrane fusion glycoprotein in cell entry, and neuraminidase (NA), which functions as the receptor destroying enzyme in virus release, form the basis for defining subtypes [3]. To date, 16 HA (H1–H16) and 9 NA (N1–N9) have been identified in avian species [4], while in the last century, only three subtypes, H1N1 in 1918 and 2009, H2N2 in 1957, and H3N2 in 1968 [5], [6], [7], have successfully adapted to humans. Hemagglutinin binds to sialic acid (SA) glycans present on host cell surfaces. The receptors on epithelial cells of the human upper respiratory tract are mainly α2-6-linked SA moieties [8]. Since avian influenza viruses predominately bind α2-3-linked SA, and human influenza viruses preferentially bind to α2-6-linked SA, human infection by avian influenza viruses is rare [9]. However, since 1997 a growing number of human cases of avian influenza infection have been reported [10], including H5N1, H7N2, H7N3, H7N7, and H9N2 strains [11]. Although the current situation with the pandemic H1N1 influenza virus dominates public health efforts, the prospect of a novel pandemic emerging from these isolated cases continues to be a major public health threat around the world.
Early cases of human infection by H7 influenza viruses are reported as far back as 1979 [12], [13]. Since 2002, multiple outbreaks and human infections of H7 subtype viruses; within both Eurasian and North American lineages have been reported. In the Netherlands in 2003, a highly pathogenic avian influenza (HPAI) H7N7 outbreak resulted in more than 80 cases of human infections, including one fatality [14], [15]. In New York in 2003, a single case of human respiratory infection of H7N2 was reported [16] and in British Columbia in 2004, an H7N3 virus caused two cases of conjunctivitis [17], [18]. More recently in 2007, the United Kingdom reported several cases of low pathogenic avian influenza (LPAI) H7N2 virus infections that caused influenza-like illness and conjunctivitis [19].
Since 1996, H7 viruses of the North American lineage have been circulating in regional live bird markets [20], containing a 24-nucleotide deletion resulting in an eight amino acid deletion in the receptor-binding site (RBS) of HA (Figure S1). The recent human infections with H7 in North America have raised public health concerns as to how these viruses adapt to such a dramatic structural change while remaining one of the predominant circulating viral strains. A recent study of H7 viruses isolated from previous outbreaks revealed efficient replication in both mouse and ferret animal models [21]. In particular, ferret studies with A/New York/107/2003 (NY107), an H7N2 virus isolated from a man in New York, not only showed efficient replication in the upper respiratory tract of the ferret but also the capacity for intra-species transmission by direct contact [21], [22]. Interestingly, both an increased preference for α2-6 and decreased preference for α2-3-linked sialosides of this virus compared to the other avian influenza viruses was shown by previous glycan microarray analysis but less so by a competitive solid-phase binding assay [22], [23].
Here we report a detailed molecular analysis of the RBS of the HA from North American lineage H7N2 virus, NY107, including glycan microarray analyses and structural analyses of the HA in complex with an avian receptor analog (3′-Sialyl-N-acetyllactosamine, 3′SLN) and two human receptor analogs (6′-Sialyl-N-acetyllactosamine, 6′SLN; Sialyllacto-N-tetraose b, LSTb). These results provide important insight into the interaction of H7 HAs with both avian and human hosts.
Results
Overall structure
By using x-ray crystallography, the structure of H7 HA from the NY107 virus was determined to 2.6 Å resolution (Table 1). In addition, we also report three H7 HA receptor complex structures, with avian receptor analog (3′SLN) to 2.7 Å resolution and with human receptor analogs (6′SLN and LSTb) to 3.0 Å and 2.6 Å resolution, respectively (Table 1). The overall structure of NY107 is similar to other reported HA structures with a globular head containing the RBS and vestigial esterase domain, and a membrane proximal domain with its distinctive, central helical stalk and HA1/HA2 cleavage site (Figure 1A). Although five asparagine-linked glycosylation sites are predicted in the NY107 HA monomer, interpretable electron density was observed at only two sites, Asn38 in HA1 and Asn82 in HA2 (all residue numbers are based on H3 numbering). At these sites, only one or two N-acetyl glucosamines could be interpreted.
Fig. 1. NY107 HA monomer and comparison of its RBS to other HA structures. 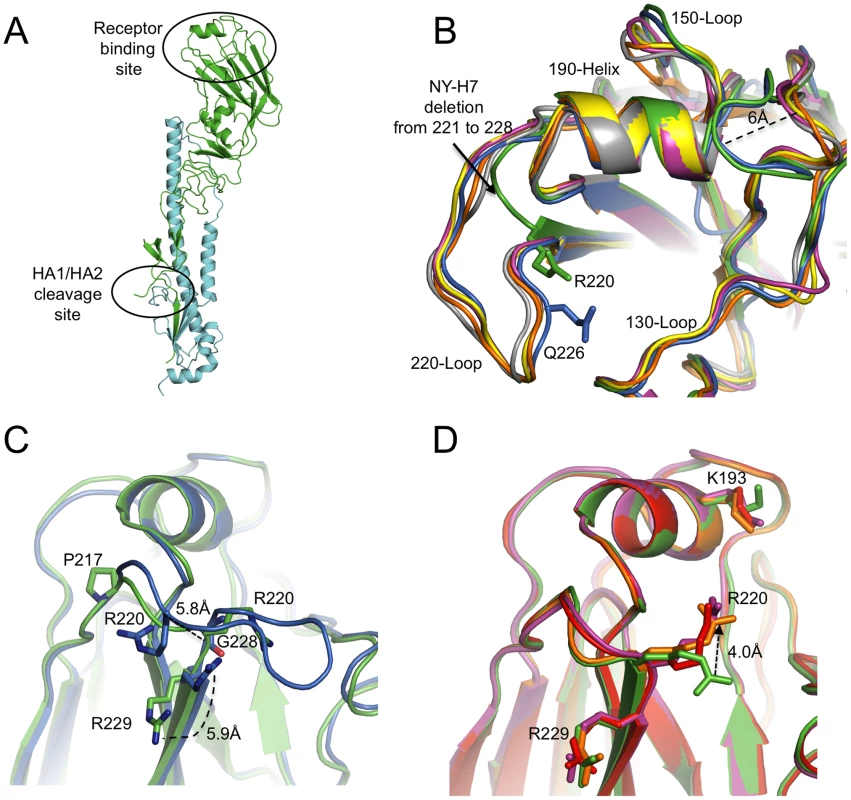
(A) One monomer is shown with the HA1 chain colored in green and the HA2 chain in cyan. The location of the receptor binding site and the HA1/HA2 cleavage site are circled. (B) The superposition of receptor binding domains of NY107 (green), Av-H7 (marine), 1918-Hu-H1 (magenta), Hu-H5 (yellow), Hu-H3 (orange), and Sw-H9 (grey). The proximity of Arg220 and Gln226 are highlighted. Three structural elements comprising this binding site are labeled. The two major differences are the extended 150-loop and the deletion of 220-loop of NY107. (C) Overlap of NY107 (green) and Av-H7 (marine) (PDB: 1TI8) illustrates the compensatory effect of R220 bringing it close to the position occupied by G228 in the avian HA. (D) Overlap of the NY107 (green), NY107- 3′SLN (orange), NY107-6′SLN (red), and NY107-LSTb (magenta) structures. All the figures were generated and rendered with the use of MacPyMOL [56]. Tab. 1. Data collection and refinement statistics. 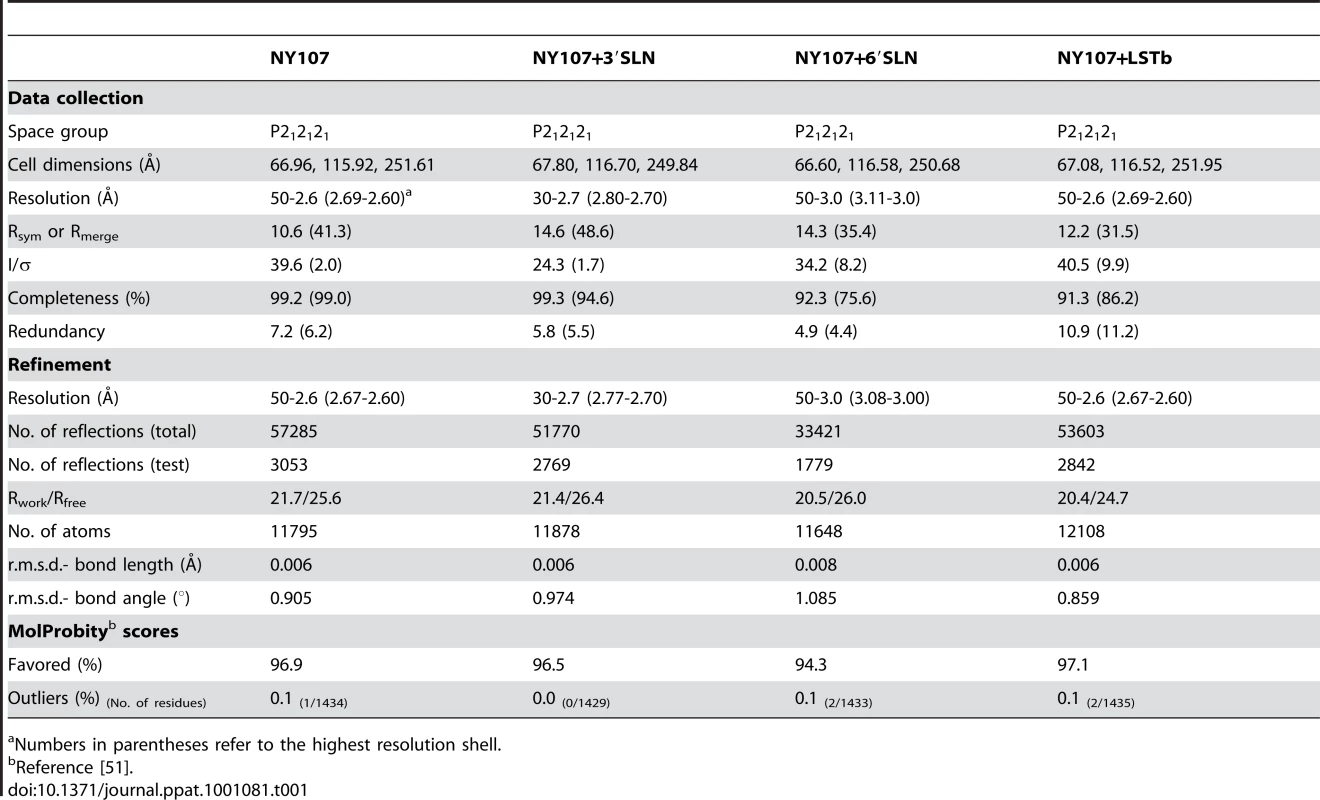
Numbers in parentheses refer to the highest resolution shell. During viral replication, HA is synthesized as a single chain precursor (HA0) and cleaved by specific host proteases into the infectious HA1/HA2 form. In baculovirus expression systems, highly pathogenic HAs, with a polybasic cleavage site, are expressed as an HA1/HA2 form [24], whereas HAs with monobasic cleavage sites (single Arg) from low pathogenic viruses are expressed as the HA0 form [25]. NY107 is regarded as a low pathogenic virus, and as expected, was produced in the HA0 form (Figure S2). However, subsequent digestion with thrombin protease to remove the His-tag resulted in cleavage to a profile on SDS-PAGE comparable to that of an HA1/HA2 form (Figure S2). A comparison of the NY107 cleavage site with the consensus cleavage pattern in the MEROPS database (http://merops.sanger.ac.uk) suggests it to be a possible thrombin cleavage site.
Based on their molecular phylogenies, HAs are divided into two groups and five clades: group 1 includes H8, H9, and H12; H1, H2, H5, and H6; H11, H13 and H16; group 2 includes H3, H4, and H14; H7, H10 and H15 [26]. Among all available HA structures, we selected ten representative HAs from both avian and human subtypes for structural analysis. As expected, NY107 HA is structurally very similar to the Avian-H7 in all comparisons and closely related to H3, the other group 2 members used in the analyses (Tables S1 and S2).
The receptor binding site
The RBS is at the membrane distal end of each HA monomer and its specificity for sialic acid and the nature of its linkage to a vicinal galactose residue is a major determinant of host range-restriction. The consensus RBS for all current HAs is composed of three major structural elements: a 190-helix (residues 188–194), a 220-loop (residues 221–228), and a 130-loop (residues 134–138). In addition, highly conserved residues (Tyr98, Trp153, His183, and Tyr195) form the base of the pocket.
Although the NY107 RBS is similar to other subtypes (H1, H2, H3, H5, and H9), a previously observed specific feature of H7 HAs, is also observed in the NY107 150-loop region: two residues inserted at position 158 result in this loop protruding more than 6Å towards the binding site compared to other subtype HAs (Figure 1B and Table S2) [27]. More interestingly, the eight amino acid deletion, only found in the North American lineage H7s, from position 221 to 228 (Figure S1), resulted in a complete loss of the 220-loop (Figure 1B). Sequence alignment shows that Arg220 and Arg229 are conserved in all influenza A HA subtypes (Figure S1), but structural alignment of NY107 HA shows Arg220 occupying the Gly228 position, and the much shorter loop turns at residue Pro217 (Figure 1C). The Cα distance between NY107 Arg220 and its homolog in the Av-H7 structure (PDB: 1TI8) [27] is 5.8Å, and they point in opposite directions (Figure 1C). The side chain direction of Av-H7 Arg220 is almost parallel with the beta sheet after Arg229, whereas the NY107 Arg220 points downward to the binding pocket. The Cα position of Arg229 in both H7 structures remains the same, except the side chain in the NY107 swings away by about 5.9Å (Figure 1C) and could help to stabilize this region by forming a hydrogen bond to the mainchain carbonyl of Gln210 in the neighboring monomer. In the absence of the 220-loop in NY107 HA, upon glycan binding the long side chain of Arg220 compensates for its loss and is displaced 4Å upward to form hydrogen bonds with receptor analogs inside the binding pocket (Figure 1D).
Effect of loop truncation on the receptor binding specificity of NY107
Previously, mutations in the HA receptor binding domains of H1N1 (Glu190Asp/Gly225Asp) and H2N2/H3N2 (Gln226Leu and Gly228Ser) subtypes were responsible for adaptation of these viruses to pandemic strains [24], [28], [29], [30]. Due to missing residues 221–228 in the NY107 HA RBS, neither mechanism for adaptation is possible. Thus, in order to look more closely at the role of the missing loop and its effect on receptor specificity, we first subjected the recombinant HA (recHA) to glycan microarray analyses and compared it to a reverse genetics-derived NY107 virus, and a co-circulating Eurasian virus and recHA, A/Netherlands/219/2003 (NL219), that has the consensus avian sequence in the 220-loop and it also infected a human [15].
Glycan microarray analysis of recombinant NY107 (Figure 2A and Table 2) revealed a highly restricted binding profile with strong binding to only α2-3 sulfated (#4–8), α2-3 branched (#9–11) and mixed α2-3/α2-6 branched sialosides (#60–64) as well as to the long linear sialyl di - and tri-lactosamines (#22, 24). Weak binding was also observed (above background) to other α2-3 glycans on the array. The recombinant NY107 also revealed a strict glycan binding preference to only one α2-6 glycan, the internal structure, Galβ1-3(Neu5Acα2-6)GlcNAcβ1-3Galβ1-4Glc (#58; LSTb) (Figure 2A), a glycan highlighted in a previous study [22]. The virus with higher valency and avidity revealed stronger binding to all α2-3 groups, in addition to the branched di-sialyl α2-6 biantennary structures (#46–48) as well the LSTb (#58) (Figure 2B and Table 2). In contrast, the NL219 recHA (Figure 2C and Table 2) bound well to only the avian α2-3 containing sialyl-glycans (sulfated, branched, linear and fucosylated). Its corresponding virus also reflected this specificity although it also revealed strong binding to α2-3 N-glycolylneuraminic acid (Neu5Gc) containing glycans (#66–70) (Figure 2D and Table 2).
Fig. 2. Receptor specificity of NY107 recHA and virus. 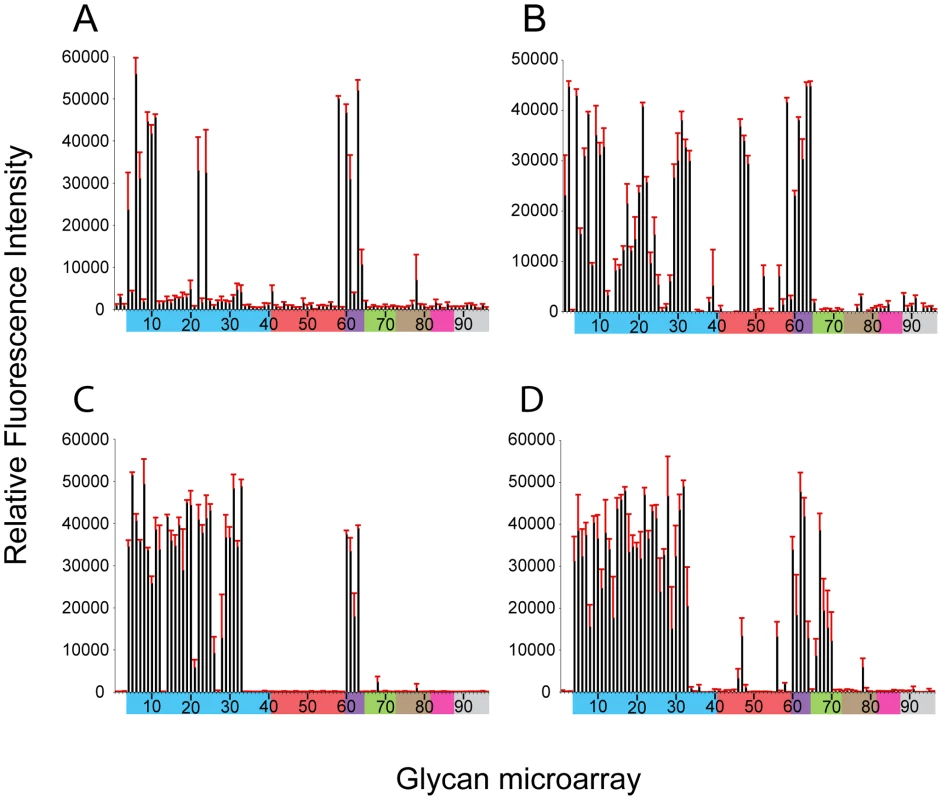
Glycan microarray analysis of recombinant NY107 HA (A) and NY107 virus (B) compared to the recHA (C) and virus (D) from a Eurasian lineage A/Netherlands/219/2003 H7 influenza virus that was circulating in the same year and also infected a human. Colored bars highlight glycans that contain α2-3 SA (blue) and α2-6 SA (red), α2-6/α2-3 mixed SA (purple), N-glycolyl SA (green), α2-8 SA (brown), β2-6 and 9-O-acetyl SA, and non-SA (grey). Error bars reflect the standard error in the signal for six independent replicates on the array. Structures of each of the numbered glycans are found in Table S4. Tab. 2. Comparison of the sialoside receptor specificity of the HAs from H7 influenza viruses. 
Members of each group are identified according to the graph number used in the microarray data in Figures 2 and 3 and correspond to numbers in the complete glycan list (Table S4). To further assess the effect of the missing 220 loop on HA structural stability and receptor specificity it was essential to evaluate these functions on the ancestral HA containing the full length 220-loop. To this end, we engineered an HA with an avian H7 consensus (PQVNGQSG) 220-loop re-introduced (NY107-220ins) into the NY107 HA and recovered this virus by reverse genetics. Compared to the NY107 virus (Figure 2A) glycan microarray analyses of the resulting NY107-220ins virus (Figure 3A and Table 2) revealed a decrease in binding to branched (#9–11) and linear (#12–27) α2-3 sialosides and a loss of binding to the branched di-sialyl α2-6 biantennary structures (#46–48), LSTb (#58) as well as the mixed α2-3/α2-6 branched sialosides (#60–64). In addition, sequence analysis of the NY107-220ins HA revealed the presence of quasispecies in the second position of the inserted loop, P(Q/K)VNGQSG, suggesting that re-introduction of the loop alone is not tolerated and does not create an avian-type binding profile. Thus other amino acid substitutions in the HA might have co-evolved with the deletion of the 220 loop to help stabilize the RBS/HA to maintain functionality.
Fig. 3. Effect of 220-loop deletion and additional RBS mutations on NY107 receptor specificity. 
NY107 was engineered to restore the 220-loop to a consensus full-length HA from 1996 (A) and additional co-variant amino acid substitutions, Glu186Gly (B), Arg205Gly (C) and the double mutant Glu186Gly/Arg205Gly (D) to restore, on the NY107 framework, an HA RBS found in viruses prior to the introduction of the deletion in North American viruses. Colored bars group glycans as described in Figure 3. Error bars reflect the standard error in the signal for six independent replicates on the array. Structures of each of the numbered glycans are found in Table S4. When viruses containing this 220-loop deletion emerged in North America in the mid 90's, four additional amino acid substitutions, Gly114Arg, Asp119Gly, Gly186Glu and Gly205Arg, in the HA1 as well as an Asp19Asn in the HA2 chain were also introduced to most of the circulating isolates. Of these, Gly186Glu and Gly205Arg in the HA1 are close to the RBS, at the monomer interface, and could potentially modulate its structure and/or function. NY107 viruses with a restored consensus 220-loop and a single Glu186Gly (NY107-ins-186) or Arg205Gly (NY107-ins-205) substitution as well as the Glu186Gly/Arg205Gly double substitution (NY107-ins-186/205) were derived by reverse genetics and evaluated. Glycan microarray analysis for the three resulting viruses revealed similar glycan binding profiles with increased binding to α2-3 sialosides, including mixed α2-3/α2-6 branched sialosides (#60–64), α2-3 Neu5Gc (#66–70), but limited binding to the α2,6 sialosides (Figures 3B, 3C, 3D), resulting in a binding profile virtually identical to that of the NL219 virus and other avian influenza viruses (Figure 2D) [30]. Sequence analysis of the three reverse genetics derived viruses revealed no mutations/quasispecies in the HAs of either the NY107-ins-186 or the NY107-ins-186/205 virus stocks, indicative of replication fitness. For the NY107-ins-205 virus however, a Glu186Gly substitution emerged in the HA after only two passages in eggs following recovery from DNA transfection, indicating the importance of the co-variant position 186 with respect to HA functionality/glycan specificity. Altogether, the data indicates that the H7 subtype avian influenza viruses that were circulating in aquatic birds and poultry in North America before 1996 exhibited a classic avian α2-3 sialoside binding preference. In order for the 220-loop deletion to be tolerated, concurrent Gly186Glu and Gly205Arg substitutions in the vicinity of RBS of HA emerged to achieve a restricted α2-3 binding profile and only a moderate/limited increase in binding to branched di-sialyl α2-6 biantennary structures (#46–48) as well the α2,6 internal sialoside, LSTb (#58).
NY107 avian receptor complex
To understand from a structural perspective how NY107 interacts with host receptors, we solved the structure of NY107 in complex with an avian and two human receptor analogs. For the avian receptor analog, 3′SLN, the electron density maps revealed well-ordered features for the Sia-1, Gal-2, and GlcNAc-3 in the NY107 HA complex structure (Figure 4A). Structural comparison of NY107 HA binding to other, H1, H2, H3, H5, and H9 subtypes (Figure S2A) revealed that 3′SLN binding to NY107 resembled binding of the other published HAs. Indeed, the terminal Sia-1 moiety is positioned almost identically in all structures, and forms the majority of hydrogen bonds and contacts with residues in the RBS (Figure 4A and Table S3).
Fig. 4. Glycan interactions within the NY107 RBS. 
The top panel shows the interactions of NY107 with (A) 3′SLN, (B) 6′SLN and (C) LSTb. NY107 is shown in orange/red/magenta cartoon respectively. The interacting HA residues are shown as green sticks. The bottom panel shows the electron density map of the ligands. The NY107 is shown in the same colors as above, and the ligands are shown as green sticks, the 2fo-fc electron density maps (contoured at 1σ) are shown in grey. Simulated annealing omit maps are shown in supplementary Figure S4. Published avian HA structures with an intact 220-loop form very close interactions with Gal-2 of 3′SLN via residue Gln226 which is important in receptor specificity and host adaptation. For example, in the avian H7/3′SLN HA structure it interacts with Gal-2 O4 [31]. In the NY107 HA structure, although Gln226 is absent and no other residue occupies the same space as Gln226 (Figure 1B), Arg220 does forms a hydrogen bond between Arg220 NH2 and Gal-2 O4 (Figure 4A). Interestingly, although there was interpretable density for the GlcNAc-3 (Figure 4A and Figure S4B), no hydrogen bonding was apparent between the HA and the GlcNAc-3, which is consistent with other reported structures [32]. Thus, for binding to avian receptors, the trans conformation of α2-3 linkages is essential and perhaps only the first two saccharides are required. Indeed, due to the absence of 220-loop in the NY107 HA structure, the “aperture” of the RBS formed by 220-loop and 130-loop in regular HAs is increased by ∼10 Å, so that the branched, internal, and perhaps more complicated glycans might be accommodated more efficiently.
NY107 human receptor complexes
In the NY107/6′SLN complex, only Sia-1 and Gal-2 are ordered (Figure 4B). The Sia-1 remains in the same position as previously analyzed glycan/HA complexes from H1, H2, H3, H5, and H9 (Figure S3B), whereas the Av-H7 complex structure with Sialyllacto-N-tetraose c (LSTc) did not reveal any density for the Sia-1 in the receptor binding site [31]. The Gal-2 position varies significantly among different subtypes. Compared to the human-adapted H1 HA [32], Gal-2 in the NY107 HA is 3Å higher, and thus is further from the protein (Figure S3B). In NY107, the Gal-2 only forms an intramolecular, saccharide-saccharide interaction with Sia-1. The poor electron density map and fewer interactions with protein residues suggest that the cis conformation of α2-6 linkages in 6′SLN trisaccharides show a reduced binding affinity with NY107.
Glycan array results with NY107 revealed a strong binding signal for the internal α2-6 sialoside, LSTb. To further investigate this interaction, we solved the structure of the NY107/LSTb complex. The final model contained Sia-1, NAG-2, Gal-3, and Gal-5 in the RBS. Although glycan microarray data indicated NY107 to have a specific affinity for LSTb, few interactions were apparent from the crystal structure. Sia-1 still forms multiple hydrogen bonds with residues in the RBS (Table S3 & Figure 4C). The branched Gal-5 interacts with Ser137, to help stabilize the LSTb binding. However, Arg220 and Lys193, the two residues showing close binding with 3′SLN, did not form any hydrogen bonds with LSTb. In the structure, Gal-5 also interacts with a crystal packing symmetry mate and thus the flexibility of whole LSTb may be restricted. In solution, with more freedom, the LSTb should be able to tilt closer to the RBS, and thus Glc-4 may have more interactions with the 190-helix than seen in the crystal structure.
Discussion
Human infections by avian influenza viruses, including H7 subtypes, continue to pose a major public health threat. Although the species barrier prevents avian influenza viruses from widespread infection of the human population, the molecular determinants of efficient interspecies transmission and pathogenicity are still poorly understood. The viral coat protein HA however, is perhaps a critical molecule since previous pandemic viruses modified their receptor specificity and overcame the interspecies barrier to spread in the human population. Although HA structures alone and in complex with receptor analogs provide considerable insight into receptor binding, it is clear that HAs from different species and subtypes have significant structural variation. Indeed, low-pathogenic H7N2 avian influenza viruses with an 8 amino acid deletion within its RBS started to circulate in live-bird markets in the northeast United States in 1996. Despite what one would consider a debilitating mutation, these viruses have been reported as the predominant isolate [33]. Whether such a deletion contributed to their evolutionary success and how are an important questions, especially in light of NY107's ability to produce respiratory illness in humans [16], as well as its reported increased affinity for human-type receptors and ability for contact transmission in ferrets [21]. To try to help answer these questions, we have analyzed the molecular structures of NY107 and its complexes with receptor analogs to explain receptor specificity at the molecular level.
The crystal structures of NY107 and its complexes with both avian and human receptor analogs describe a mechanism as to how an influenza virus might adapt by dramatically altering its RBS, and still be functional. Arg220 of the HA1 chain of NY107 compensates for the loss of the 220-loop, by forming hydrogen bonds with Gal-2 from the avian analog (binding was not observed in either of the structures complexes with the human analogs). However, in the LSTb complex, branched Gal-5 forms extra interactions with the 130-loop, thus improving the binding preference for this particular glycan. Consistent with the structural evidence, glycan microarray analyses of NY107 revealed a strong binding preference for the branched α2-6 sialoside, LSTb. Except for the absence of the 220-loop, other key residues within the RBS are conserved in NY107 and thus, direct interactions with sialic acid are maintained.
The 220-loop is recognized as one of the three crucial structural elements in the RBS. Aside from the North American lineage H7N2 viruses, which have been circulating with a deletion (221–228) in this loop, there has been one other report describing a seven amino acid deletion (224–230) in a laboratory generated H3N2 escape mutant which was reported to have a slightly increased affinity for α2-3-linked glycans by hemagglutination assay [34]. Meanwhile, the equivalent region in the hemagglutinin-esterase-fusion (HEF) protein of influenza C virus reveals a rearrangement resulting in a truncated 260-loop in its RBS (Figure S5) [35]. However, without structural data with appropriate receptor analogs, it is not possible to compare the role of these loop variants in receptor binding to the H7 HA structure described here.
When compared to NL219, another co-circulating H7 avian virus HA (Figure 2C and D), overall binding to α2-3-linked glycans was markedly reduced, while increased binding to α2-6-linked receptors was only marginal. However, these results focus attention on only 2 sub-classes of human-type receptors that may be important for infection (and transmission in ferrets). The NY107 virus interaction with biantennary glycans (Figure 2B), although weak (not seen in Figure 2A with recHA), is a possible route for virus entry as biantennary structures are common on tissues, i.e. glycan profiling data from human lung tissue on the Consortium for Functional Glycomics (CFG) web site. In addition, the internal sialoside, LSTb, was observed in both virus and recHA microarray data, suggesting this type of glycan has good affinity for this HA. The significance of this is unknown since LSTb has only been described in human milk [36].
Interestingly, NY107 and NL219 virus receptor binding and specificity has been addressed previously using glycan microarray analysis that reported a significantly increased preference for α2-6 and decreased preference for α2-3-linked sialosides [22]. In addition, the same viruses were also included in a recent study from Gambaryan et al. using a competitive solid-phase binding assay [23]. Our findings confirm and extend the receptor binding specificity reported by these authors in that they reported both viruses binding to sulfated sialylglycans with a lactosamine (Galβ1-4GlcNAc core and reported only a moderate binding affinity for α2-6-sialyllactosamine, the human-type receptor analog used in their assay.
The 220-loop is an integral feature of the receptor binding site, and thus one would predict that such a deletion might have compromised this strain to be deleted from the population of circulating viruses. However, this was not the case [33] and its existence appears to be in part due to the additional mutations at positions 186 and 205. Restoration of the loop with either or both residues mutated back to the pre-1994 consensus sequence resulted in a classic avian influenza virus binding profile. The emergence of the Glu186Gly mutation in the HA of the NY107-ins-205 mutant after only two passages of the rescued virus in eggs, also indicates the importance of these positions for HA functionality/glycan specificity. Analysis of the structural data reveals that positions 186 and 205 are on opposite sides of a monomer but are both close to the 220-loop deletion region in the trimeric form. The Glu at position 186 is close to Arg220 and may interact with Arg220 when binding avian receptors. Position 205 in the neighboring monomer may be important in trimer stability and maintaining RBS functionality. If one models the pre-1996 220-loop restored into the NY107 structure, Arg205, Glu186 and the loop all clash, thus explaining the Glu186Gly mutation that emerged in the NY107-ins-205 virus HA after limited egg passage.
The NY107 RBS with its more restricted α2-3 glycan binding preference and weak/moderate increase in α2-6 binding may have given the virus a selective advantage to be maintained in poultry at live bird markets and supplying farms. Certain terrestrial birds, such as quails and chickens, have recently been shown to present both human and avian types of receptors in the trachea and intestine [37], [38], [39]. Although it is not known what specific glycans are presented in these animals, it is conceivable that a virus with mixed specificity might have a distinct advantage over avian viruses that have specific avian receptor requirements, particularly in bird markets where multiple species coalesce. Previous results with H7N2, H9N2 and H5N1 viruses all highlight the fact that an increase in α2-6-binding preference is not sufficient for efficient transmission of avian influenza viruses to humans [22], [40], [41]. Although it remains to be seen whether prolonged circulation of viruses in terrestrial birds, such as domestic chickens, can provide a possible route for viruses to adapt for efficient human infection [11], continued surveillance of influenza viruses from avian and other animal reservoirs is urgently needed to define their zoonotic potential.
Materials and Methods
Cloning
Based on H3 numbering [42], cDNA corresponding to residues 11–329 (HA1) and 1–176 (HA2) of the ectodomain of the hemagglutinin (HA) from A/New York/107/2003 (H7N2; Genbank:ACC55270) and A/Netherlands/219/2003 (H7N7; Genebank: AAR02640) was cloned into the baculovirus transfer vector, pAcGP67-A (BD Biosciences), incorporating a C-terminal thrombin cleavage site, a “foldon” sequence [43] and a His-tag at the extreme C-terminus of the construct to enable protein purification [25], [44]. Transfection and virus amplification were carried out according to the baculovirus expression system manual (BD Biosciences Pharmingen).
Protein expression and purification
Soluble NY107 was recovered from the cell supernatant by metal affinity chromatography using Ni-NTA resin (Qiagen Inc.). Fractions containing NY107 were pooled and dialyzed against 10 mM Tris-HCl, 50 mM NaCl, pH 8.0, then subjected to ion-exchange chromatography (IEX) using a Mono-Q HR 10/10 column (GE Healthcare). IEX purified NY107 was subjected to thrombin digest (3 units/mg protein; overnight at 4°C) and purified by gel filtration chromatography using a Superdex-200 16/60 column (GE Healthcare) and 50 mM Tris-HCl, 100 mM NaCl, pH 8.0 as running buffer. Protein eluting as a trimer was buffer exchanged into 10 mM Tris-HCl, 50 mM NaCl, pH 8.0 and concentrated to 14.5 mg/ml for crystallization trials. At this stage, the protein sample still contained the additional plasmid-encoded residues at both the N (ADPG) and C terminus (SGRLVPR).
Crystallization, ligand soaking and data collection
Initial crystallization trials were set up using a Topaz Free Interface Diffusion (FID) Crystallizer system (Fluidigm Corporation, San Francisco, CA). Crystals were observed in several conditions containing PEG 3350 or PEG 4000. Following optimization, diffraction quality crystals for NY107 were obtained at room temperature using a modified method for microbath under oil [45], by mixing the protein with reservoir solution containing 20% PEG 3350, 0.2 M magnesium chloride at pH 7.2. For receptor analog complexes, crystals were soaked for 3 hours in the crystallization buffer containing 10 mM 3′SLN or 6′SLN (V-labs Inc., Covington, LA), or overnight in 10mM LSTb (Sigma, St. Louis, MO). All crystals were flash-cooled at 100K using 20% glycerol as the cryo-protectant. Datasets were collected at Advanced Photon Source (APS) beamlines 22 ID and BM at 100K. Data were processed with the DENZO-SACLEPACK suite [46]. Statistics for data collection are presented in Table 1.
Structure determination and refinement
The structure of NY107 was determined by molecular replacement with Phaser [47] using the structure of the avian H7 (Av-H7) from A/turkey/Italy/2002, pdb:1TI8 (HA1, 78% identity; HA2, 90% identity) as the searching model. One HA trimer occupies the asymmetric unit with an estimated solvent content of 58% based on a Matthews' coefficient (Vm) of 2.9 Å3/Da. Rigid body refinement of the trimer led to an overall R/Rfree of 28.6%/37.4%. The model was then “mutated” to the correct sequence and rebuilt by Coot [48], then the protein structures were refined with REFMAC [49] using TLS refinement [50]. The final models were assessed using MolProbity [51]. The three complex structures were refined and evaluated using the same strategy. All statistics for data processing and refinement are presented in Table 1. Electron density maps (2fo-fc) were generated in Refmac [49] while simulated annealing omit maps were generated by sa-omit-map, a part of the Crystallography and NMR System (CNS) software [52].
Virus generation
Wild type and mutant viruses of NY107 (H7N2) and A/Netherland/219/2003 (H7N7) were generated from plasmids by a reverse genetics approach [53]. To generate viruses with amino acid insertion or substitution in the HA, mutations were introduced into plasmid DNA with an overlap extension PCR approach [54]. Viruses derived by plasmid transfection of HK293 cells were propagated in eggs. The genomes of resulting virus stocks were sequenced to detect the emergence of possible variants during amplification.
Glycan binding analyses
Glycan microarray printing and recHA analyses have been described previously [24], [30], [44], [55] (see Table 2 for glycans used for analyses in these experiments). Virus were analyzed on the microarray as described previously [30].
PDB accession codes
The atomic coordinates and structure factors of NY107 are available from the RCSB PDB under accession codes 3M5G for the unliganded NY107, 3M5H for the NY107 with 3′-SLN and 3M5I and 3M5J for NY107 with 6′SLN and LSTb, respectively.
Accession/ID numbers for genes/proteins used in this work
A/New York/107/03 (H7N2), Genbank: ACC55270; A/Netherlands/219/03 (H7N7), Genbank: AAR02640; A/Hong Kong/1-9/68 (H3N2), 2HMG; A/Duck/Ukraine/1/63 (H3N8), PDB: 1MQL; A/South Carolina/1/18 (H1N1), PDB: 1RD8; A/Puerto Rico/8/34 (H1N1), PDB: 1RU7; A/Swine/Iowa/15/30 (H1N1), PDB: 1RUY; A/Singapore/1/1957 (H2N2), PDB: 2WRC; A/Viet Nam/1203/04 (H5N1), PDB: 2FK0; A/Duck/Singapore/3/97 (H5N3), PDB: 1JSM; A/Swine/Hong Kong/9/98 (H9N2), PDB: 1JSD; A/Turkey/Italy/8000/02 (H7N3), PDB: 1TI8; C/Johannesburg/1/66, 1FLC.
Supporting Information
Zdroje
1. ThompsonWW
ShayDK
WeintraubE
BrammerL
CoxN
2003 Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289 179 186
2. ThompsonWW
ShayDK
WeintraubE
BrammerL
BridgesCB
2004 Influenza-associated hospitalizations in the United States. JAMA 292 1333 1340
3. WHO 1980 A revision of the system of nomenclature for influenza viruses: a WHO memorandum. Bull, WHO 58 585 591
4. FouchierRA
MunsterV
WallenstenA
BestebroerTM
HerfstS
2005 Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol 79 2814 2822
5. ScholtissekC
RohdeW
Von HoyningenV
RottR
1978 On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology 87 13 20
6. KawaokaY
BeanWJ
WebsterRG
1989 Evolution of the hemagglutinin of equine H3 influenza viruses. Virology 169 283 292
7. GartenRJ
DavisCT
RussellCA
ShuB
LindstromS
2009 Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325 197 201
8. ShinyaK
EbinaM
YamadaS
OnoM
KasaiN
2006 Avian flu: influenza virus receptors in the human airway. Nature 440 435 436
9. MatrosovichMN
GambaryanAS
TenebergS
PiskarevVE
YamnikovaSS
1997 Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 233 224 234
10. de JongJC
ClaasEC
OsterhausAD
WebsterRG
LimWL
1997 A pandemic warning? Nature 389 554
11. TaubenbergerJK
MorensDM
FauciAS
2007 The next influenza pandemic: can it be predicted? JAMA 297 2025 2027
12. WebsterRG
GeraciJ
PeturssonG
SkirnissonK
1981 Conjunctivitis in human beings caused by influenza A virus of seals. N Engl J Med 304 911
13. KurtzJ
ManvellRJ
BanksJ
1996 Avian influenza virus isolated from a woman with conjunctivitis. Lancet 348 901 902
14. KoopmansM
WilbrinkB
ConynM
NatropG
van der NatH
2004 Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 363 587 593
15. FouchierRA
SchneebergerPM
RozendaalFW
BroekmanJM
KeminkSA
2004 Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A 101 1356 1361
16. CDC 2004 Update: influenza activity–United States and worldwide, 2003–04 season, and composition of the 2004–05 influenza vaccine. 547 552 MMWR Morb Mortal Wkly Rep: Centers for Disease Control
17. HirstM
AstellCR
GriffithM
CoughlinSM
MoksaM
2004 Novel avian influenza H7N3 strain outbreak, British Columbia. Emerg Infect Dis 10 2192 2195
18. TweedSA
SkowronskiDM
DavidST
LarderA
PetricM
2004 Human illness from avian influenza H7N3, British Columbia. Emerg Infect Dis 10 2196 2199
19. EditorialTeam 2007 Avian influenza A/H7N2 outbreak in the United Kingdom. Euro Surveill 12 2
20. SuarezDL
GarciaM
LatimerJ
SenneD
PerdueM
1999 Phylogenetic analysis of H7 avian influenza viruses isolated from the live bird markets of the Northeast United States. J Virol 73 3567 3573
21. BelserJA
LuX
MainesTR
SmithC
LiY
2007 Pathogenesis of avian influenza (H7) virus infection in mice and ferrets: enhanced virulence of Eurasian H7N7 viruses isolated from humans. J Virol 81 11139 11147
22. BelserJA
BlixtO
ChenLM
PappasC
MainesTR
2008 Contemporary North American influenza H7 viruses possess human receptor specificity: Implications for virus transmissibility. Proc Natl Acad Sci U S A 105 7558 7563
23. GambaryanAS
TuzikovAB
PazyninaGV
DeshevaJA
BovinNV
2008 6-sulfo sialyl Lewis X is the common receptor determinant recognized by H5, H6, H7 and H9 influenza viruses of terrestrial poultry. Virol J 5 85
24. StevensJ
BlixtO
GlaserL
TaubenbergerJK
PaleseP
2006 Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol 355 1143 1155
25. StevensJ
CorperAL
BaslerCF
TaubenbergerJK
PaleseP
2004 Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303 1866 1870
26. RussellRJ
KerryPS
StevensDJ
SteinhauerDA
MartinSR
2008 Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc Natl Acad Sci U S A 105 17736 17741
27. RussellRJ
GamblinSJ
HaireLF
StevensDJ
XiaoB
2004 H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology 325 287 296
28. MatrosovichM
TuzikovA
BovinN
GambaryanA
KlimovA
2000 Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol 74 8502 8512
29. NobusawaE
IshiharaH
MorishitaT
SatoK
NakajimaK
2000 Change in receptor-binding specificity of recent human influenza A viruses (H3N2): a single amino acid change in hemagglutinin altered its recognition of sialyloligosaccharides. Virology 278 587 596
30. StevensJ
BlixtO
ChenLM
DonisRO
PaulsonJC
2008 Recent avian H5N1 viruses exhibit increased propensity for acquiring human receptor specificity. J Mol Biol 381 1382 1394
31. RussellRJ
StevensDJ
HaireLF
GamblinSJ
SkehelJJ
2006 Avian and human receptor binding by hemagglutinins of influenza A viruses. Glycoconj J 23 85 92
32. GamblinSJ
HaireLF
RussellRJ
StevensDJ
XiaoB
2004 The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303 1838 1842
33. SuarezDL
SpackmanE
SenneDA
2003 Update on molecular epidemiology of H1, H5, and H7 influenza virus infections in poultry in North America. Avian Dis 47 888 897
34. DanielsPS
JeffriesS
YatesP
SchildGC
RogersGN
1987 The receptor-binding and membrane-fusion properties of influenza virus variants selected using anti-haemagglutinin monoclonal antibodies. EMBO J 6 1459 1465
35. RosenthalPB
ZhangX
FormanowskiF
FitzW
WongCH
1998 Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature 396 92 96
36. WeinsteinJ
de Souza-e-SilvaU
PaulsonJC
1982 Purification of a Gal beta 1 to 4GlcNAc alpha 2 to 6 sialyltransferase and a Gal beta 1 to 3(4)GlcNAc alpha 2 to 3 sialyltransferase to homogeneity from rat liver. J Biol Chem 257 13835 13844
37. GambaryanA
WebsterR
MatrosovichM
2002 Differences between influenza virus receptors on target cells of duck and chicken. Arch Virol 147 1197 1208
38. WanH
PerezDR
2006 Quail carry sialic acid receptors compatible with binding of avian and human influenza viruses. Virology 346 278 286
39. GuoCT
TakahashiN
YagiH
KatoK
TakahashiT
2007 The quail and chicken intestine have sialyl-galactose sugar chains responsible for the binding of influenza A viruses to human type receptors. Glycobiology 17 713 724
40. MainesTR
ChenLM
MatsuokaY
ChenH
RoweT
2006 Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A 103 12121 12126
41. WanH
SorrellEM
SongH
HossainMJ
Ramirez-NietoG
2008 Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS One 3 e2923
42. WeisWI
BrungerAT
SkehelJJ
WileyDC
1990 Refinement of the influenza virus hemagglutinin by simulated annealing. J Mol Biol 212 737 761
43. FrankS
KammererRA
MechlingD
SchulthessT
LandwehrR
2001 Stabilization of short collagen-like triple helices by protein engineering. J Mol Biol 308 1081 1089
44. StevensJ
BlixtO
TumpeyTM
TaubenbergerJK
PaulsonJC
2006 Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312 404 410
45. ChayenNE
Shaw-StewardPD
BlowDM
1992 Microbatch crystallization under oil – a new technique allowing many small volume crystallization experiments. J Cryst Growth 122 176 180
46. OtwinowskiA
MinorW
1997 Processing of X-ray diffraction data collected in oscillation mode. Meothds in Enzymology 276 307 326
47. McCoyAJ
Grosse-KunstleveRW
StoroniLC
ReadRJ
2005 Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61 458 464
48. EmsleyP
CowtanK
2004 Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 2126 2132
49. CCP4 1994 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50 760 763
50. WinnMD
IsupovMN
MurshudovGN
2001 Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57 122 133
51. DavisIW
Leaver-FayA
ChenVB
BlockJN
KapralGJ
2007 MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35 W375 383
52. BrungerAT
2007 Version 1.2 of the Crystallography and NMR system. Nat Protoc 2 2728 2733
53. HoffmannE
WebsterRG
2000 Unidirectional RNA polymerase I-polymerase II transcription system for the generation of influenza A virus from eight plasmids. J Gen Virol 81 2843 2847
54. HiguchiR
KrummelB
SaikiRK
1988 A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res 16 7351 7367
55. BlixtO
HeadS
MondalaT
ScanlanC
HuflejtME
2004 Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A 101 17033 17038
56. DeLanoWL
2002 The PyMol Molecular Graphics Systems. wwwpymolorg
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek SRFR1 Negatively Regulates Plant NB-LRR Resistance Protein Accumulation to Prevent AutoimmunityČlánek Inhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on VirulenceČlánek Phylogenetic Approach Reveals That Virus Genotype Largely Determines HIV Set-Point Viral LoadČlánek A Family of Plasmodesmal Proteins with Receptor-Like Properties for Plant Viral Movement Proteins
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 9- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Azole Drugs Are Imported By Facilitated Diffusion in and Other Pathogenic Fungi
- Two Genes on A/J Chromosome 18 Are Associated with Susceptibility to Infection by Combined Microarray and QTL Analyses
- Impact of Simian Immunodeficiency Virus Infection on Chimpanzee Population Dynamics
- Breaking the Stereotype: Virulence Factor–Mediated Protection of Host Cells in Bacterial Pathogenesis
- The Canine Papillomavirus and Gamma HPV E7 Proteins Use an Alternative Domain to Bind and Destabilize the Retinoblastoma Protein
- Rescue of HIV-1 Release by Targeting Widely Divergent NEDD4-Type Ubiquitin Ligases and Isolated Catalytic HECT Domains to Gag
- Steric Shielding of Surface Epitopes and Impaired Immune Recognition Induced by the Ebola Virus Glycoprotein
- Dynamics of the Multiplicity of Cellular Infection in a Plant Virus
- HLA Class I Binding of HBZ Determines Outcome in HTLV-1 Infection
- Pathogenic Bacteria Target NEDD8-Conjugated Cullins to Hijack Host-Cell Signaling Pathways
- The HA and NS Genes of Human H5N1 Influenza A Virus Contribute to High Virulence in Ferrets
- SRFR1 Negatively Regulates Plant NB-LRR Resistance Protein Accumulation to Prevent Autoimmunity
- Cyclin-Dependent Kinase Activity Controls the Onset of the HCMV Lytic Cycle
- The N-Terminal Domain of the Arenavirus L Protein Is an RNA Endonuclease Essential in mRNA Transcription
- Generation of Neutralizing Antibodies and Divergence of SIVmac239 in Cynomolgus Macaques Following Short-Term Early Antiretroviral Therapy
- Inhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on Virulence
- Intracellular Proton Conductance of the Hepatitis C Virus p7 Protein and Its Contribution to Infectious Virus Production
- The Transcriptome of the Human Pathogen at Single-Nucleotide Resolution
- The Epidermal Growth Factor Receptor (EGFR) Promotes Uptake of Influenza A Viruses (IAV) into Host Cells
- Surface Co-Expression of Two Different PfEMP1 Antigens on Single -Infected Erythrocytes Facilitates Binding to ICAM1 and PECAM1
- Sequestration and Tissue Accumulation of Human Malaria Parasites: Can We Learn Anything from Rodent Models of Malaria?
- Phylogenomics of Ligand-Gated Ion Channels Predicts Monepantel Effect
- Generation of Covalently Closed Circular DNA of Hepatitis B Viruses via Intracellular Recycling Is Regulated in a Virus Specific Manner
- CpG-Methylation Regulates a Class of Epstein-Barr Virus Promoters
- Molecular and Evolutionary Bases of Within-Patient Genotypic and Phenotypic Diversity in Extraintestinal Infections
- A Bistable Switch and Anatomical Site Control Virulence Gene Expression in the Intestine
- Are Members of the Fungal Genus (a) Commensals; (b) Opportunists; (c) Pathogens; or (d) All of the Above?
- Structures of Receptor Complexes of a North American H7N2 Influenza Hemagglutinin with a Loop Deletion in the Receptor Binding Site
- Phylogenetic Approach Reveals That Virus Genotype Largely Determines HIV Set-Point Viral Load
- The Coevolution of Virulence: Tolerance in Perspective
- Involvement of the Cytokine MIF in the Snail Host Immune Response to the Parasite
- Structure of the Extracellular Portion of CD46 Provides Insights into Its Interactions with Complement Proteins and Pathogens
- A Family of Plasmodesmal Proteins with Receptor-Like Properties for Plant Viral Movement Proteins
- High Content Phenotypic Cell-Based Visual Screen Identifies Acyltrehalose-Containing Glycolipids Involved in Phagosome Remodeling
- A Novel Small Molecule Inhibitor of Hepatitis C Virus Entry
- The Microbiota Mediates Pathogen Clearance from the Gut Lumen after Non-Typhoidal Diarrhea
- RNA Polymerases (L-Protein) Have an N-Terminal, Influenza-Like Endonuclease Domain, Essential for Viral Cap-Dependent Transcription
- Pathogen Specific, IRF3-Dependent Signaling and Innate Resistance to Human Kidney Infection
- Cellular Entry of Ebola Virus Involves Uptake by a Macropinocytosis-Like Mechanism and Subsequent Trafficking through Early and Late Endosomes
- The Length of Vesicular Stomatitis Virus Particles Dictates a Need for Actin Assembly during Clathrin-Dependent Endocytosis
- Formation of Mobile Chromatin-Associated Nuclear Foci Containing HIV-1 Vpr and VPRBP Is Critical for the Induction of G2 Cell Cycle Arrest
- Association of Tat with Promoters of PTEN and PP2A Subunits Is Key to Transcriptional Activation of Apoptotic Pathways in HIV-Infected CD4+ T Cells
- Metal Hyperaccumulation Armors Plants against Disease
- Cyclin-Dependent Kinase-Like Function Is Shared by the Beta- and Gamma- Subset of the Conserved Herpesvirus Protein Kinases
- Role of Acetyl-Phosphate in Activation of the Rrp2-RpoN-RpoS Pathway in
- Ebolavirus Is Internalized into Host Cells Macropinocytosis in a Viral Glycoprotein-Dependent Manner
- A Novel Family of IMC Proteins Displays a Hierarchical Organization and Functions in Coordinating Parasite Division
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Structure of the Extracellular Portion of CD46 Provides Insights into Its Interactions with Complement Proteins and Pathogens
- The Length of Vesicular Stomatitis Virus Particles Dictates a Need for Actin Assembly during Clathrin-Dependent Endocytosis
- Inhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on Virulence
- The Coevolution of Virulence: Tolerance in Perspective
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



