-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaAssociation of the Polymorphism His615Arg with Melanin Content in East Asian Populations: Further Evidence of Convergent Evolution of Skin Pigmentation
The last decade has witnessed important advances in our understanding of the genetics of pigmentation in European populations, but very little is known about the genes involved in skin pigmentation variation in East Asian populations. Here, we present the results of a study evaluating the association of 10 Single Nucleotide Polymorphisms (SNPs) located within 5 pigmentation candidate genes (OCA2, DCT, ADAM17, ADAMTS20, and TYRP1) with skin pigmentation measured quantitatively in a sample of individuals of East Asian ancestry living in Canada. We show that the non-synonymous polymorphism rs1800414 (His615Arg) located within the OCA2 gene is significantly associated with skin pigmentation in this sample. We replicated this result in an independent sample of Chinese individuals of Han ancestry. This polymorphism is characterized by a derived allele that is present at a high frequency in East Asian populations, but is absent in other population groups. In both samples, individuals with the derived G allele, which codes for the amino acid arginine, show lower melanin levels than those with the ancestral A allele, which codes for the amino acid histidine. An analysis of this non-synonymous polymorphism using several programs to predict potential functional effects provides additional support for the role of this SNP in skin pigmentation variation in East Asian populations. Our results are consistent with previous research indicating that evolution to lightly-pigmented skin occurred, at least in part, independently in Europe and East Asia.
Published in the journal: . PLoS Genet 6(3): e32767. doi:10.1371/journal.pgen.1000867
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000867Summary
The last decade has witnessed important advances in our understanding of the genetics of pigmentation in European populations, but very little is known about the genes involved in skin pigmentation variation in East Asian populations. Here, we present the results of a study evaluating the association of 10 Single Nucleotide Polymorphisms (SNPs) located within 5 pigmentation candidate genes (OCA2, DCT, ADAM17, ADAMTS20, and TYRP1) with skin pigmentation measured quantitatively in a sample of individuals of East Asian ancestry living in Canada. We show that the non-synonymous polymorphism rs1800414 (His615Arg) located within the OCA2 gene is significantly associated with skin pigmentation in this sample. We replicated this result in an independent sample of Chinese individuals of Han ancestry. This polymorphism is characterized by a derived allele that is present at a high frequency in East Asian populations, but is absent in other population groups. In both samples, individuals with the derived G allele, which codes for the amino acid arginine, show lower melanin levels than those with the ancestral A allele, which codes for the amino acid histidine. An analysis of this non-synonymous polymorphism using several programs to predict potential functional effects provides additional support for the role of this SNP in skin pigmentation variation in East Asian populations. Our results are consistent with previous research indicating that evolution to lightly-pigmented skin occurred, at least in part, independently in Europe and East Asia.
Introduction
The remarkable variation observed in skin, hair and iris pigmentation in human populations is the result of differences in the amount, type and distribution of the pigment melanin, which is synthesized by specialized cells known as melanocytes. Pigmentation is a complex trait, influenced by numerous genes and their interactions. The last decade has witnessed impressive advances in our understanding of the genetics of normal pigmentation variation, driven by functional studies, gene expression studies, studies in animal models, analyses of signatures of natural selection and candidate gene or genome-wide association studies. At least 11 genes are known to be associated with normal pigmentation variation: TYR, TYRP1, OCA2/HERC2, SLC45A2, SLC24A5, SLC24A4, MC1R, ASIP, KITLG, IRF4 and TPCN2 [Reviewed in 1],[2]. In European and related populations, a clear picture is emerging of the genetic and evolutionary processes associated with skin lightening. The most important genes involved are SLC24A5, SLC45A2 and KITLG, which explain a large portion of the skin pigmentation differences observed between European and West African populations [3]–[6]. Other polymorphisms within the genes TYR, OCA2, MC1R, ASIP and IRF4 are known to play a role in normal pigmentation variation in populations of European descent [7]–[13]. A recent genome-wide study reported that variants within the genes SLC24A5, SLC45A2 and TYR are associated with skin pigmentation in a South Asian sample [14]. Interestingly, the genes SLC24A5 and SLC45A2 show an extremely unusual pattern of allele frequency distribution, with a derived allele near fixation in European populations, and the alternative ancestral allele fixed in other population groups [5]. These two genes show strong signatures of selection in European populations [15]–[19], but not in other population groups. These observations indicate that evolution to lightly-pigmented skin happened, at least in part, independently in Europe and East Asia [5],[16]. Unfortunately, while there have been important advances in our understanding of the genetics of pigmentation in European populations, very little is known about the genes involved in skin pigmentation variation in East Asian populations. There are also pigmentation candidate genes (DCT, ADAM17, ADAMTS20 , KITLG, TYRP1 and OCA2) that show signatures of selection in East Asians [16]–[21], but formal association studies are required to confirm that these genes are involved in skin pigmentation variation. Studies of signatures of natural selection are extremely useful as a strategy to identify potential genes of interest, but it is critical to carry out further analysis in order to confirm that the signatures of selection are not false positives and to eliminate the possibility that positive selection was related to biological processes other than pigmentation [22],[23]. For example, the ADAM17 gene has been implicated in many processes involved in cell-cell and cell-matrix interactions, including fertilization, muscle development and neurogenesis. Therefore, it is in principle possible that the signatures of selection observed in this gene are due to its role in these processes.
Here, we present the results of a study evaluating the association of 10 Single Nucleotide Polymorphisms (SNPs) located within 5 pigmentation candidate genes (OCA2, DCT, ADAM17, ADAMTS20 and TYRP1) with skin pigmentation measured quantitatively in a sample of individuals of East Asian ancestry living in Canada. We selected these loci based on previous studies that identified signatures of natural selection in East Asian populations, and prioritized a list of SNPs within these genes using the program SNPSelector. We show that the non-synonymous polymorphism rs1800414 located within the OCA2 gene is significantly associated with skin pigmentation in this East Asian sample. We replicated this result in an independent sample of Chinese individuals of Han ancestry. This polymorphism is characterized by a derived allele that is present at high frequency in East Asian populations, but is absent in other populations. In our sample, individuals with the derived G allele, which codes for the amino acid arginine, show lower melanin levels than those with the ancestral A allele, which codes for the amino acid histidine. An analysis of this non-synonymous polymorphism using several programs to predict potential functional effects (see materials and methods) provides additional support for the role of this SNP in skin pigmentation variation in East Asian populations.
Results
We applied four tests of positive selection based on different statistics to the five genes analyzed in this study. For these tests, we used genomewide information available for the HapMap East Asian, European and African samples (see Material and methods section). Table 1 shows the results of the four tests of natural selection in the HapMap sample. In accordance to previous reports [16]–[21], we observed evidence of positive selection in East Asian populations for these pigmentation genes. The OCA2 gene shows numerous SNPs displaying high levels of differentiation in the East Asian sample with respect to the genomewide average (LSBL tests), very negative Tajima's D values for two windows encompassing a portion of this gene and a reduction in genetic diversity. We also observed clusters of markers exhibiting high differentiation for the DCT gene, as well as evidence of a reduction of genetic diversity in the East Asian sample for this locus (lnRH test). The ADAM17 gene is significant for the LSBL, lnRH and Tajima's D tests, and is also significant for the WGLRH test, indicating that ADAM17 has haplotypes characterized by derived alleles that have risen to very high frequencies and have longer than expected levels of Linkage Disequilibrium (LD). The gene ADAMTS20 has extreme values for the LSBL and Tajima's D statistics. Finally, markers in the gene TYRP1 show high levels of genetic differentiation between East Asians and the other two HapMap populations measured by LSBL and this locus is encompassed by a significant extended haplotype region (WGLRH test).
Tab. 1. Tests of positive selection for the five pigmentation genes analyzed in this study in the East Asian HapMap sample. 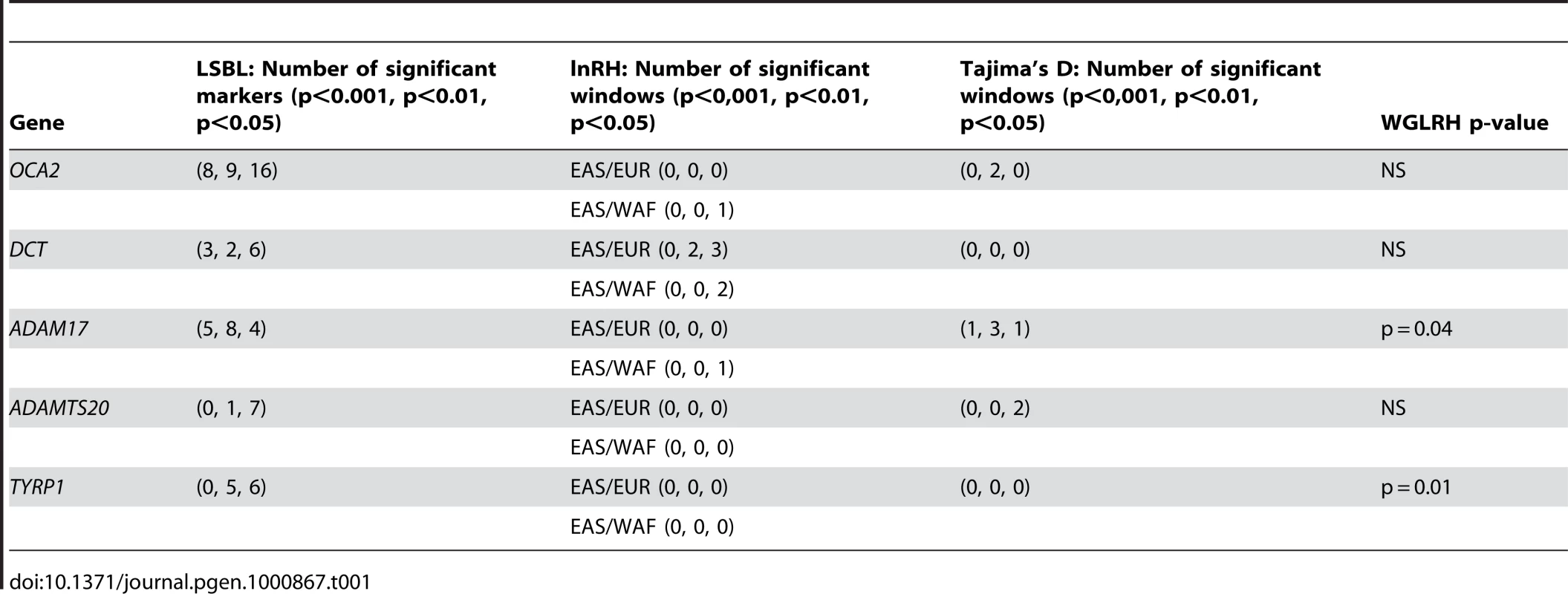
Ten polymorphisms located within these five genes were genotyped in a sample of individuals of East Asian ancestry (N = 122). Table 2 reports the genotype and allele frequencies for each marker. No significant deviations from Hardy-Weinberg proportions were identified for any of the SNPs. We evaluated the patterns of LD between the markers located in each gene using an Expectation Maximization algorithm implemented in the program EMLD. LD was low between the markers located within the OCA2 gene (rs7495174/rs1800414: r2 = 0.06; rs7495174/rs1545397: r2 = 0.05 and rs1800414/rs1545397: r2 = 0.25). In contrast, there was perfect LD between the markers located within the DCT gene (rs1407995/rs2031526: r2 = 1). Finally, within the ADAMTS20 gene, there was almost perfect LD between the markers rs11182091 and rs11182085 (r2>0.99), but LD was substantially lower between rs11182091 and rs1510523 (r2 = 0.30) and rs11182085 and rs1510523 (r2 = 0.31).
Tab. 2. Observed and expected genotype frequencies, allele frequencies, and the Hardy-Weinberg exact test for 10 SNPs in the Canadian East Asian sample. 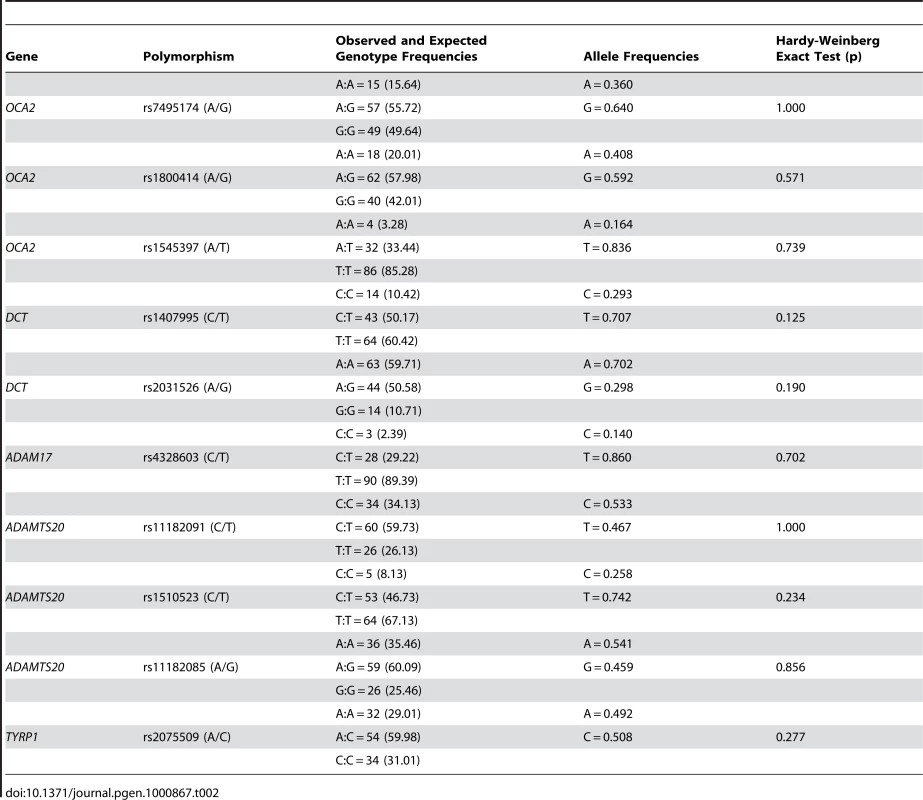
We tested if there was evidence of association between the 10 SNPs and quantitative measures of constitutive pigmentation (melanin index) in the East Asian sample. The results of the linear regression analysis for each marker, including sex as a covariate are depicted in Table 3. The rs1800414 polymorphism located within the OCA2 gene showed a significant association with skin pigmentation. Using an additive model, we estimated that each copy of the G allele decreases skin pigmentation by approximately 1.3 melanin units (p = 0.002) and the rs1800414 polymorphism explains approximately 9% of the pigmentation variation observed in this sample. A model-free (unconstricted) analysis indicates that AG heterozygotes decrease skin pigmentation by 1.6 melanin units (p = 0.046) and GG homozygotes by 2.6 melanin units (p = 0.002), with respect to AA homozygotes. The marker rs1800414 remains significant when using the conservative Bonferroni correction (taking into account the intermarker LD patterns and assuming 8 independent tests, the p-value after correction is p = 0.016). Figure 1 shows the distribution of melanin index value by rs1800414 genotype. No association was observed for the other 9 SNPs analyzed in this study.
Fig. 1. Boxplot showing melanin index by rs1800414 genotype for the Canadian East Asian sample. 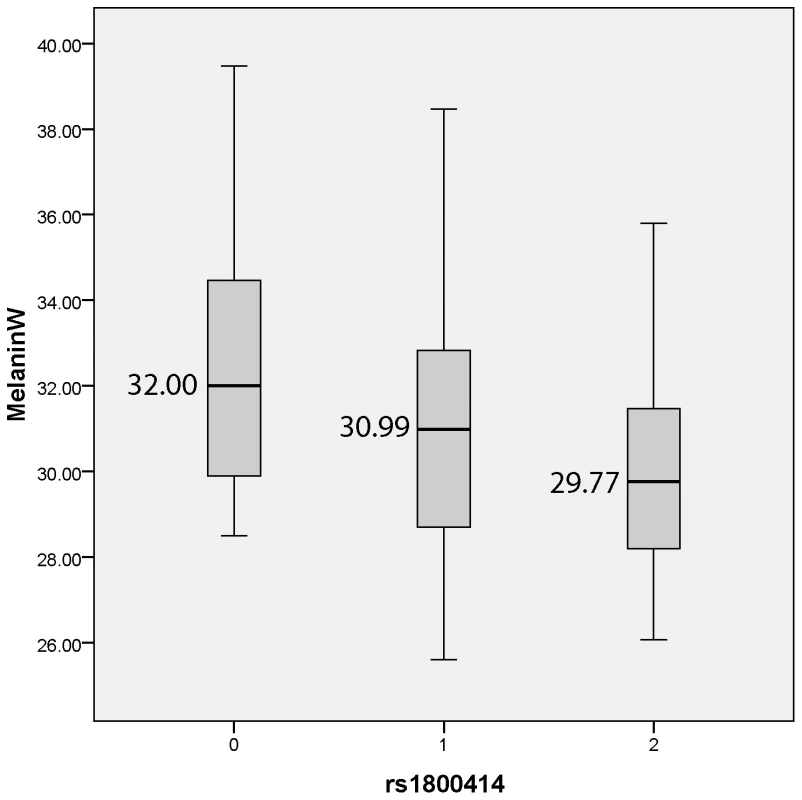
The top of the box is the 75th percentile, the bottom of the box is the 25th percentile and the line in the centre is the median. The lines extending from the box mark the highest and lowest melanin index measurements. Tab. 3. Linear regression coefficients and p-values for each of the 10 SNPs in the Canadian East Asian sample. 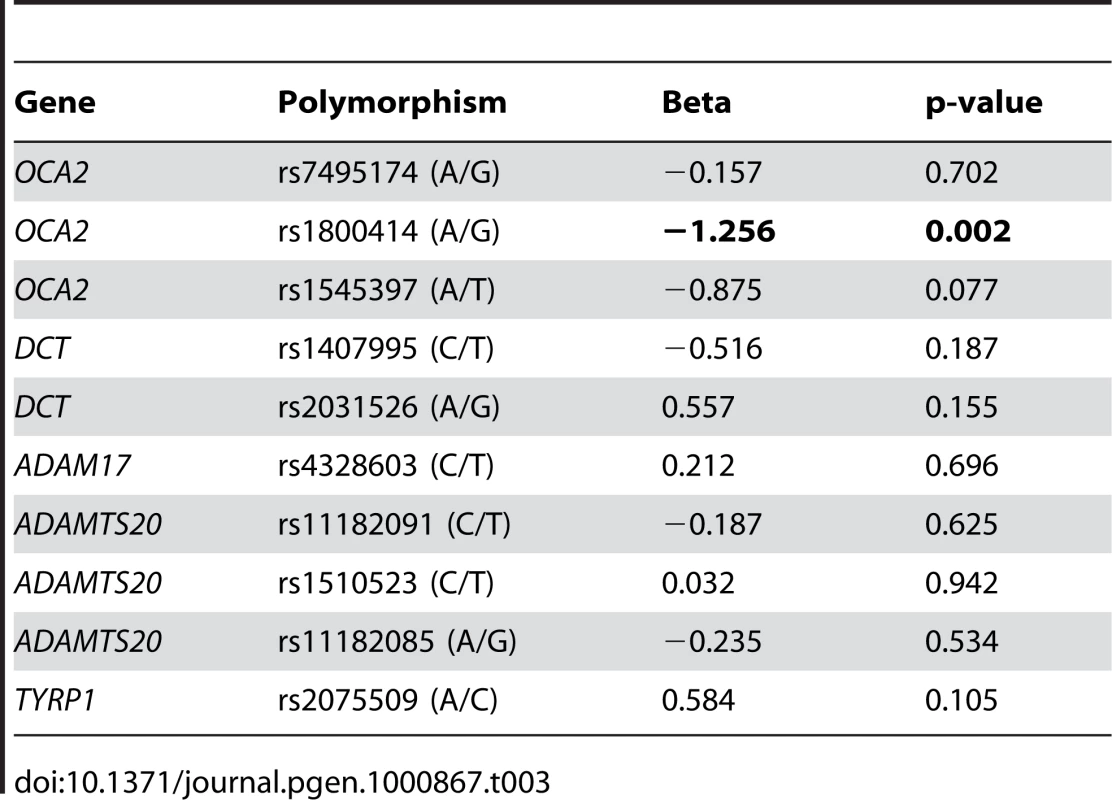
In order to confirm the results of this study, we genotyped the rs1800414 polymorphism in an independent sample of Chinese individuals of Han ancestry recruited in China (N = 207) and measured with a similar instrument. The results of this analysis are reported in Table 4. In agreement with our preliminary results, the linear regression analysis shows that rs1800414 has a significant effect on skin pigmentation, although the effect size is slightly lower than in our study. Under an additive model, each copy of the G allele decreases skin pigmentation by 0.85 melanin units (p = 0.005) and explains around 4% of the variation observed in the sample. Under a model-free (unconstrained) model, the AG heterozygotes decrease skin pigmentation by approximately 1 melanin unit (p = 0.085) and the GG homozygotes by 1.7 melanin units (p = 0.005), with respect to AA homozygotes.
Tab. 4. Observed and expected genotype frequencies, allele frequencies, and Hardy-Weinberg exact test for rs1800414 in the Chinese Han sample. 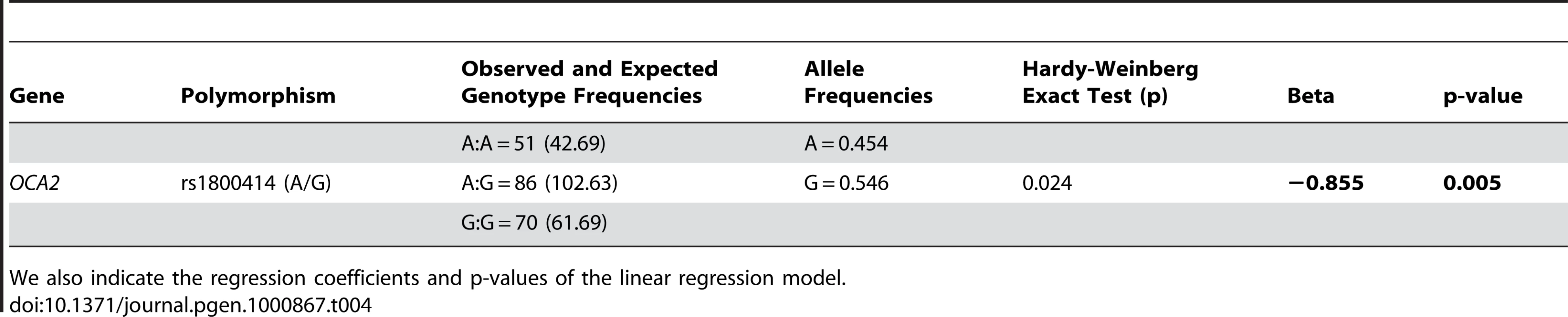
We also indicate the regression coefficients and p-values of the linear regression model. Discussion
We analyzed the association of 10 SNPs within 5 pigmentation candidate genes (OCA2, DCT, ADAM17, ADAMTS20 and TYRP1) with skin melanin content measured quantitatively in an East Asian sample. Previous studies have indicated that these 5 genes show signatures of natural selection in East Asian populations [16]–[20] and our analysis of signatures of selection using data obtained with the Affymetrix 6.0 chip showed a remarkable agreement with these studies. The 10 SNPs selected for analysis showed high allele frequency differences between East Asian and non-Asian populations and 6 of them (rs1800414, rs7495174, rs1182091, rs1510523, rs11182085 and rs2075509) also had high function, regulatory or phastcons scores in SNPSelector, which indicated that these SNPs could be of functional importance. We observed that one of the markers included in the study, the non-synonymous SNP rs1800414 (His615Arg) located within the OCA2 gene, was significantly associated with melanin index in our sample of Canadian individuals of East Asian ancestry (p = 0.002). An analysis in an independent sample of Chinese individuals of Han ancestry also showed that the His615Arg polymorphism has a significant effect on skin pigmentation (p = 0.005). Based on both samples, it can be estimated that each copy of the derived G allele (coding for the amino acid Arginine), which is present at high frequency in East Asian populations, but absent in European and West African populations, decreases skin pigmentation by 0.85–1.3 melanin units. Additionally, the unconstrained statistical analysis shows that in terms of its effects on skin pigmentation, this polymorphism fits a codominant model of inheritance, rather than dominant or recessive models. Although significant, the phenotypic effect observed for rs1800414 is lower than the effect that has been reported for other polymorphisms previously associated with skin pigmentation. For example, studies in African American populations have shown that polymorphisms located within the pigmentation genes SLC24A5, SLC45A2 and KITLG have an effect of more than 3 melanin units per allele copy [3],[5],[6]. However, direct comparison between studies is complicated by the different pigmentation characteristics of the samples. This is one of the first formal reports of association with skin pigmentation measured using reflectometry in East Asian populations. Our study indicates that the OCA2 gene was independently involved in the evolution of light pigmentation in Europe and East Asia, and in combination with previous findings for other genes (SLC24A5 and SLC45A2), strongly suggests that there was convergent evolution towards light pigmentation in Europe and East Asia. Previous studies reported that the OCA2/HERC2 gene showed distinct signatures of positive selection in Europe and East Asia [16],[19],[20],[21]. Markers in the HERC2 gene are associated with blue eyes in European and related populations. In particular, the SNP rs12913832 segregates almost perfectly with blue-brown eye color [24]–[26]. This SNP is located within a highly conserved region that may act as a control region for OCA2 and a recent study reported that rs12913832 had a significant effect on the levels of OCA2 mRNA [25],[27]. Lao et al. [19] reported that the OCA2 gene had significant Extended Haplotype Homozygosity (EHH) values in European and East Asian samples, but the core haplotypes were different in both populations. Yuasa et al. [28] noted that the rs1800414 G allele (R615) is very frequent in East Asian populations, but rare or absent in African and Indo-European populations. Anno et al. [29] also showed that European and East Asian populations are characterized by different haplotypes at the OCA2 gene, with the East Asian haplotype harboring the variant rs1800414 G, which is the allele that is associated with light skin in our study. More recently, Donelly et al. [21] described that the rs1800414 allele is under selection in East Asia, and the blue eye allele BEH2 (defined by rs12913832) is under selection in Europe and Southwest Asia. Therefore, it seems clear that there were independent selective processes acting on the OCA2 gene in Europe and East Asia, involving distinct haplotypes.
Our sample comprises individuals of East Asian ancestry living in Toronto. The majority of the subjects have ancestry from China, South Korea and Japan (N = 96), but some individuals have ancestry from Southeast Asia (Vietnam, Thailand and Phillipines, N = 26). If there are large differences in frequency between East Asian populations for rs1800414, our significant association results for this SNP could be confounded by population stratification. However, two observations indicate that this is not the case: 1/ There are no significant deviations from Hardy-Weinberg proportions for any of the markers included in our study (Table 2). The effect of allele frequency differences between East Asian populations would have been reflected in deviations from Hardy-Weinberg (excess of homozygotes, Wahlund effect). In fact, there is a slight excess of heterozygotes for rs1800414 in our total sample, which is the opposite of what would be expected in the presence of stratification, 2/ The statistical analysis excluding the Southeast Asian subjects (N = 96) is also significant and shows remarkable concordance with the results obtained using the full sample (beta = −1.6, p = 0.001). In this respect, it is important to note that Yuasa [28] reported that there are no large frequency differences between five samples from China and Japan for the rs1800414 G allele (44.8%–63%). Similarly, we did not observe significant allele frequency differences between the Canadian East Asian sample and the Chinese sample that was used for replication (p = 0.255).
Our statistical analysis was significant for rs1800414, but not for the other SNPs genotyped in this sample, including 2 additional SNPs within the OCA2 gene. Given our relatively small sample size, our study was not adequately powered to identify loci with small effects. The rs1800414 polymorphism explains a substantial proportion of the skin pigmentation variation observed in the sample. A relevant question is if the observed effects are due to rs1800414 or to a causative SNP in LD with rs1800414 within the OCA2 gene. In this sense, there is strong evidence pointing to rs1800414 as the causative variant itself. In addition to SNPSelector, we used other tools to infer the functional effect of this polymorphism (FastSNP, the SNP function portal, SIFT and Polyphen). All of these methods suggest that this non-synonymous rs1800414 SNP, which was first described by Lee et al. [30], is functionally important. FastSNP indicates that the functional effect of this SNP may be mediated through the regulation of alternative splicing. The programs Polyphen and SIFT also point to a damaging effect of the A to G transition at rs1800414. It would be extremely important to carry out gene expression studies of this polymorphism, similar to the research published for other variants known to be associated with skin pigmentation using primary cultures of human melanocytes [2],[27],[31].
Our study provides new evidence regarding the genetic and evolutionary processes driving the lightening of skin following the migration of anatomically modern humans from Africa to high latitude regions in Europe and East Asia. Evidence is growing that the reduction in melanin content took place, at least in part, independently in these two regions. We now know that the evolution of skin pigmentation has been quite complex: some genes were the target of positive selection only in one population group (eg. SLC24A5 and SLC45A2 in Europe), whereas other genes were under selection independently in more than one group (eg. OCA2 in Europe and East Asia). However, there are still many aspects of the evolution of skin pigmentation that remain unclear. Our picture of the genetics of normal pigmentation variation in non-European populations is still incomplete, and the evolutionary time frame remains to be elucidated. When did the evolution to light skin take place in Europe and East Asia? It has been suggested, based on evidence collected for the SLC24A5 gene, that the evolution to light skin occurred in Europe long after the arrival of anatomically modern humans to this continent [32], but it will be necessary to collect information on additional genes and from different geographic regions to gain a better understanding of the evolution of skin pigmentation in human populations.
Materials and Methods
Ethics statement
Written informed consent was obtained from each participant, and the study was approved by the University of Toronto Health Sciences Research Ethics Board.
Recruitment
Participants were recruited by the Molecular Anthropology Laboratory at the University of Toronto Mississauga (UTM) between 2007 and 2009. Recruitment took place primarily through the use of advertisements on UTM campus, and online advertisements in the University of Toronto community. Geographic origin was assessed using questions regarding the participant's place of birth and the ancestry of their parents and maternal and paternal grandparents. In total, 122 East Asians were recruited.
Measurement of melanin using reflectometry
We took quantitative melanin measurements from each participant's inner arm using a narrow-band reflectometer (DermaSpectrometer, Cortex Technology, Hadsund, Denmark). This instrument emits light at the green (568 nm) and red (655 nm) wavelengths of the visible spectrum and a photodetector measures the amount of light reflected by the skin. These measurements are used to estimate the melanin content in the skin, which is expressed as the Melanin Index (M). In human populations, the melanin index ranges from the low 20s (individuals with light skin) to close to 100 (individuals with dark skin). Throughout the text, when we refer to melanin units, we refer to the melanin index values obtained with the DermaSpectrometer. More information about this instrument is available in Shriver and Parra [33]. In order to capture the most accurate reading of constitutive skin pigmentation, these measurements were carried out during the winter.
Selection of SNPs in pigmentation candidate genes
We used SNPSelector [http://snpselector.duhs.duke.edu/hqsnp36.html] to prioritize a limited number of SNPs to genotype within each of the pigmentation candidate genes. SNPSelector is a SNP selection tool that provides information on population allele frequencies, linkage disequilibrium patterns, potential SNP function and patterns of SNP conservation. Our criteria for SNP selection was based on: 1/ high frequency differences between East Asian and non-Asian populations (West Africa and Europe) and 2/ potential functional effect, based on the function score, regulatory score or conservation score (PhastCons score). The following SNPs were selected for genotyping: 1/ Gene OCA2: rs1800414 is a non-synonymous polymorphism with an allele present at high frequency in East Asian populations (G allele, 59%) but absent in non-Asian populations. This SNP also had very high function, regulatory and phastcons scores; rs1545397 is an intronic polymorphism showing dramatic allele frequency differences between East Asian and non-Asian populations (>85%); rs7495174 is an intronic variant showing high frequency differences between East Asian and non-Asian populations (>45%) and a high regulatory score (CpG island), 2/ Gene DCT: rs1407995 and rs2031526 are intronic polymorphisms showing very high frequency differences between East Asian and non-Asian populations (>60%), 3/Gene ADAM17: rs4328603 is an intronic SNP showing very high frequency differences between East Asian and non-Asian populations (>60%), 4/ Gene ADAMTS20: rs11182091 is an intronic SNP with substantial frequency differences between East Asian and non-Asian populations (>30%) and a high regulatory score (conserved transcription factor binding site); rs1510523 is an intronic variant with high frequency differences between East Asian and non-Asian populations (>40%) and high regulatory and phastcons scores; rs11182085 is an intronic SNP with substantial frequency differences between East Asian and non-Asian populations (>30%) and high regulatory and phastcons scores, 5/ Gene TYRP1: rs2075509 is an intronic variant with high frequency differences between East Asian and non-Asian populations and high regulatory and phastcons scores. No markers were studied at the KITLG gene because no SNPs were identified with large frequency differences between East Asian and European populations. This is consistent with reports indicating that the signatures of selection observed in KITLG region are shared in Europeans and East Asians [6].
DNA collection and genotyping
A sample of each participant's blood was collected in a 4-mL EDTA tube. DNA was extracted from the blood using the E.Z.N.A. Blood DNA Midi Kit (Omega Bio-Tek, Georgia, United States). Genotyping was done by the company KBiosciences [http://www.kbioscience.co.uk/] using a KASPar assay that relies on competitive allele specific PCR and fluorescent detection. Eighty-nine genotypes were characterized in duplicate, and the concordance rate between the samples and the blind duplicates was 100%.
Statistical analysis
Departures from Hardy-Weinberg proportions were evaluated using an exact test available at the website http://ihg2.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl. Linkage disequilibrium (LD) between the markers located within the same genes was estimated using the program EMLD (University of Texas, Houston, TX). LD is reported as the r2 value. Association between the selected SNPs and Melanin Index was tested using linear regression. Sex was included as a covariate, as it has been found to be associated with skin pigmentation in previous studies [34],[35]. Each of the 10 SNPs was tested independently using additive and unconstrained models. The regression analysis was carried out with the program SPSS (version 17.0, SPSS Inc., 2008).
Power analysis
We used the program Quanto [http://hydra.usc.edu/gxe/] to estimate the statistical Power of our study using an additive model and a range of allele frequencies and allelic effects (measured as the regression coefficient-beta). These estimates are based on the distribution of melanin levels observed in the East Asian sample (mean melanin index = 31, standard deviation = 3) and a sample size of 120 individuals. For markers with intermediate allele frequencies (35%–65%), our study has more than 90% power to detect effects higher than 1.3 melanin units (type I error rate = 0.05, two-sided test). The Power drops for markers with more extreme frequencies: for a marker with 20% frequency, the power to identify effects higher than 1.3 is 77.8% and for a marker with 10% frequency, it is 52.5%.
Replication of significant results
For replication of the significant results of the initial analysis, the OCA2 His615Arg polymorphism was genotyped by sequencing in an independent sample from China. The sample comprised 207 individuals of Han ancestry that were recruited by Professor Li Jin at Fudan University. Skin pigmentation was measured in the inner upper arm with an instrument similar to that used to measure pigmentation in the Canadian East Asian samples (DermaSpectrometer, Cortex Technology, Hadsund, Denmark). Informed consent was obtained from each participant, and the project approved by the research ethics board of the School of Life Sciences, Fudan University.
Tests of positive selection
Four different tests of selection were used to evaluate evidence of positive selection in the HapMap East Asian sample for the five genes analyzed in this study. They include the locus-specific branch length (LSBL), the log of the ratio of heterozygosities (lnRH), Tajima's D, and whole genome long range haplotype (WGLRH) test [36]–[39]. The results reported here are based on genome-wide data for the East Asian, European and West African HapMap samples obtained with the Affymetrix 6.0 chip, which includes approximately 1 million SNPs. The LSBL test evaluates if genetic markers within a genomic region show unusual levels of differentiation with respect to the genome average. This test apportions the genetic variation observed in East Asian, European and West African populations for each SNP, and identifies markers with high levels of genetic differentiation in the East Asian sample. The lnRH test highlights genomic regions with low levels of genetic diversity in the population of interest, in comparison with other population groups. This statistic was calculated for a two-way population comparison between East Asians and Europeans, and East Asians and West Africans, using an overlapping sliding window size of 100,000 base pairs (bp) and moving in 25,000 bp increments along a chromosome. Regions of the genome with negative Tajima's D values are also a hallmark of positive selection. However, negative values of D can result from demographic events as well, specifically the recovery from a population bottleneck. For this reason, it is important to compare local values of Tajima's D with the empirical levels observed in the genome. As for the lnRH analysis, Tajima's D was calculated for each population using an overlapping sliding window size of 100,000 bp with a 25,000 bp offset. The statistical significance for each of the LSBL, lnRH, and Tajima's D statistics was based on the genome-wide empirical distribution, using the formula PE (x) = (number of loci>x)/(total number loci). The final test used to infer selection is the WGLRH test of Zhang et al. [37]. This test first calculates the Relative Extended Haplotype Homozygosity (REHH) for each core haplotype in the data set and identifies core haplotypes with longer than expected ranges of linkage disequilibrium (LD) given their frequency in the population. A gamma distribution is then estimated using maximum likelihood methods against which the REHH of each core haplotype is tested to determine if its respective p-value is suggestive of recent, positive selection. This test then considers the ancestral state of the alleles, determined by a closely related outgroup, to identify SNPs where the derived allele has risen to high frequencies (>0.60). For this data set, the ancestral state for all SNPs available in the chimpanzee sequence was retrieved using the UCSC genome browser. In total, the ancestral states for 846,032 SNPS on the autosomes and X chromosome were obtained. Lastly, the WGLRH test applies a false discovery rate approach to control for false positives and identifies significant extended haplotypes. The four statistics used in this analysis have been described in more detail in Bigham et al. [40].
Prediction of potential functional effects
We analyzed the OCA2 His615Arg polymorphism using the programs FastSNP (http://fastsnp.ibms.sinica.edu.tw/pages/input_CandidateGeneSearch.jsp), the SNP function portal (http://brainarray.mbni.med.umich.edu/Brainarray/Database/SearchSNP/snpfunc.aspx) and SIFT (http://sift.jcvi.org/) in order to predict potential functional effects.
Zdroje
1. ParraEJ
2007 Human pigmentation variation: evolution, genetic basis, and implications for public health. Am J Phys Anthropol Suppl 45 85 105
2. SturmRA
2009 Molecular genetics of human pigmentation diversity. Hum Mol Genet 18 R9 17
3. LamasonRL
MohideenMA
MestJR
WongAC
NortonHL
2005 SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310 1782 1786
4. GrafJ
HodgsonR
van DaalA
2005 Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum Mutation 25 278 284
5. NortonHL
KittlesRA
ParraE
McKeigueP
MaoX
2007 Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol 24 710 722
6. MillerCT
BelezaS
PollenAA
SchluterD
KittlesRA
2007 Cis-regulatory changes in Kit Ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell 131 1179 1189
7. ShriverMD
ParraEJ
DiosS
BonillaC
NortonH
2003 Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet 112 387 399
8. AkeyJM
WangH
XiongM
WuH
LiuW
2001 Interaction between the melanocortin-1 receptor and P genes contributes to inter-individual variation in skin pigmentation phenotypes in a Tibetan population. Hum Genet 108 516 520
9. MakovaK
NortonH
2005 Worldwide polymorphism at the MC1R and normal pigmentation variation in humans. Peptides 26 1901 1908
10. KanetskyPA
SwoyerJ
PanossianS
HolmesR
GuerryD
2002 A polymorphism in the agouti signaling protein gene is associated with human pigmentation. Am J Hum Genet 70 770 775
11. BonillaC
BoxillLA
DonaldSA
WilliamsT
SylvesterN
2005 The 8818G allele of the agouti signaling protein (ASIP) gene is ancestral and is associated with darker skin color in African Americans. Hum Genet 116 402 406
12. VoiseyJ
Gomez-Cabrera M delC
SmitDJ
LeonardJH
SturmRA
2006 A polymorphism in the agouti signaling protein (ASIP) is associated with decreased levels of mRNA. Pigment Cell Res 19 226 231
13. HanJ
KraftP
NanH
GuoQ
ChenC
2008 A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet 4 e1000074 doi:10.1371/journal.pgen.1000074
14. StokowskiRP
PantPV
DaddT
FeredayA
HindsDA
2007 A genomewide association study of skin pigmentation in a South Asian populations. Am J Hum Genet 81 1119 1132
15. VoightBF
KudaravalliS
WenX
PritchardJK
2006 A map of recent positive selection in the human genome. PLoS Biol 4 e72 doi:10.1371/journal.pbio.0040072
16. McEvoyB
BelezaS
ShriverMD
2006 The genetic architecture of normal variation in human pigmentation: an evolutionary perspective and model. Hum Mol Genet 15 Spec No 2 R176 R181
17. IzagirreN
GarciaI
JunqueraC
de la RuaC
AlonsoS
2006 A scan for signatures of positive selection in candidate loci for skin pigmentation in humans. Mol Biol Evol 23 1697 1706
18. MylesS
SomelM
TangK
KelsoJ
StonekingM
2007 Identifying genes underlying pigmentation differences among human populations. Hum Genet 120 613 621
19. LaoO
de GruijterJM
van DuijnK
NavarroA
KayserM
2007 Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann Hum Genet 71 354 369
20. AlonsoS
IzagirreN
Smith-ZubiagaI
GardeazabalJ
Diaz-RamonJL
2008 Complex signatures of selection for the melanogenic loci TYR, TYRP1 and DCT in humans. BMC Evol Biol 8 74
21. DonnellyMP
SpeedWC
KiddJR
PakstisAJ
KiddKK
2009 Selection for blue eyes in Europe and light skin pigmentation in East Asia at OCA2/HERC2. Platform Abstract #294. The American Society of Human Genetics 59th Annual meeting. Available at http://www.ashg.org/2009meeting/pdf/platforms_4up.pdf
22. KelleyJL
MadeoyJ
CalhounJC
SwansonW
AkeyJM
2006 Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res 16 980 989
23. TeshimaKM
CoopG
PrzeworskiM
2006 How reliable are empirical genomic scans for selective sweeps? Genome Res 16 702 712
24. EibergH
TroelsenJ
NielsenM
MikkelsenA
Mengel-FromJ
2008 Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum Genet 123 177 187
25. SturmRA
DuffyDL
ZhaoZZ
LeiteFP
StarkMS
2008 A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am J Hum Genet 82 424 431
26. KayserM
LiuF
JanssensAC
RivadeneiraF
LaoO
2008 Three genome-wide association studies and a linkage analysis identify HERC2 as a human iris color gene. Am J Hum Genet 82 411 423
27. CookAL
ChenW
ThurberAE
SmitDJ
SmithAG
2009 Analysis of cultured human melanocytes based on polymorphisms within the SLC45A2/MATP, SLC24A5/NCKX5, and OCA2/P loci. J Invest Dermatol 129 392 405
28. YuasaI
UmetsuK
HariharaS
KidoA
MiyoshiA
2007 Distribution of two Asian-related coding SNPs in the MC1R and OCA2 genes. Biochem Genet 45 535 542
29. AnnoS
AbeT
YamamotoT
2008 Interactions between SNP alleles at multiple loci contribute to skin color differences between Caucasoid and Mongoloid subjects. Int J Biol Sci 4 81 86
30. LeeST
NichollsRD
JongMT
FukaiK
SpritzRA
1995 Organization and sequence of the human P gene and identification of a new family of transport proteins. Genomics 26 354 363
31. SviderskayaEV
BennettDC
HoL
BailinT
LeeST
SpritzRA
1997 Complementation of hypopigmentation in p-mutant (pink-eyed dilution) mouse melanocytes by normal human P cDNA, and defective complementation by OCA2 mutant sequences. J Invest Dermatol 108 30 34
32. GibbonsA
2007 American Association of Physical Anthropologists meeting. European skin turned pale only recently, gene suggests. Science 316 364
33. ShriverMD
ParraEJ
2000 Comparison of narrow-band reflectance spectroscopy and tristimulus colorimetry for measurements of skin and hair color in persons of different biological ancestry. Am J Phys Anthropol 112 17 27
34. JablonskiNG
ChaplinG
2000 The evolution of human skin coloration. J Hum Evol 39 57 106
35. RohK
KimD
HaS
RoY
KimJ
LeeH
2001 Pigmentation in Koreans: study of the differences from caucasians in age, gender and seasonal variations. Br J Dermatol 144 94 99
36. TajimaF
1989 Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585 595
37. ZhangC
BaileyDK
AwadT
LiuG
XingG
2006 A whole genome long-range haplotype (WGLRH) test for detecting imprints of positive selection in human populations. Bioinformatics 22 2122 2128
38. ShriverM
KennedyGC
ParraEJ
LawsonHA
HuangJ
2004 The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Human Genomics 1 274 286
39. StorzJF
PayseurBA
NachmanMW
2004 Genome scans of DNA variability in humans reveal evidence for selective sweeps outside of Africa. Mol Biol Evol 21 1800 1811
40. BighamA
MaoX
BrutsaertT
WilsonM
JulianCG
2009 Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Human Genomics 4 79 90
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 3
-
Všechny články tohoto čísla
- Parental Genome Dosage Imbalance Deregulates Imprinting in
- Identification and Functional Analysis of the Vision-Specific BBS3 (ARL6) Long Isoform
- HAP2(GCS1)-Dependent Gamete Fusion Requires a Positively Charged Carboxy-Terminal Domain
- Initial Genomics of the Human Nucleolus
- Role of RecA and the SOS Response in Thymineless Death in
- PPS, a Large Multidomain Protein, Functions with Sex-Lethal to Regulate Alternative Splicing in
- Mislocalization of XPF-ERCC1 Nuclease Contributes to Reduced DNA Repair in XP-F Patients
- Transgenic Rat Model of Neurodegeneration Caused by Mutation in the Gene
- Human Population Differentiation Is Strongly Correlated with Local Recombination Rate
- Local-Scale Patterns of Genetic Variability, Outcrossing, and Spatial Structure in Natural Stands of
- Arginylation-Dependent Neural Crest Cell Migration Is Essential for Mouse Development
- HP1 Recruitment in the Absence of Argonaute Proteins in
- MiR-218 Inhibits Invasion and Metastasis of Gastric Cancer by Targeting the Robo1 Receptor
- Bias and Evolution of the Mutationally Accessible Phenotypic Space in a Developmental System
- Papillorenal Syndrome-Causing Missense Mutations in / Result in Hypomorphic Alleles in Mouse and Human
- Rapid Assessment of Genetic Ancestry in Populations of Unknown Origin by Genome-Wide Genotyping of Pooled Samples
- Regulation of Lifespan, Metabolism, and Stress Responses by the SH2B Protein, Lnk
- KRAB–Zinc Finger Proteins and KAP1 Can Mediate Long-Range Transcriptional Repression through Heterochromatin Spreading
- Identification of the Regulatory Logic Controlling Pathoadaptation by the SsrA-SsrB Two-Component System
- Drosophila Xpd Regulates Cdk7 Localization, Mitotic Kinase Activity, Spindle Dynamics, and Chromosome Segregation
- Multiple Signals Converge on a Differentiation MAPK Pathway
- In the Tradition of Science: An Interview with Victor Ambros
- Association of the Polymorphism His615Arg with Melanin Content in East Asian Populations: Further Evidence of Convergent Evolution of Skin Pigmentation
- Fatal Cardiac Arrhythmia and Long-QT Syndrome in a New Form of Congenital Generalized Lipodystrophy with Muscle Rippling (CGL4) Due to Mutations
- Deciphering Normal Blood Gene Expression Variation—The NOWAC Postgenome Study
- Derepression of the Plant Chromovirus Induces Germline Transposition in Regenerated Plants
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Deciphering Normal Blood Gene Expression Variation—The NOWAC Postgenome Study
- Transgenic Rat Model of Neurodegeneration Caused by Mutation in the Gene
- Papillorenal Syndrome-Causing Missense Mutations in / Result in Hypomorphic Alleles in Mouse and Human
- Fatal Cardiac Arrhythmia and Long-QT Syndrome in a New Form of Congenital Generalized Lipodystrophy with Muscle Rippling (CGL4) Due to Mutations
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



