-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaPatterns of Ancestry, Signatures of Natural Selection, and Genetic Association with Stature in Western African Pygmies
African Pygmy groups show a distinctive pattern of phenotypic variation, including short stature, which is thought to reflect past adaptation to a tropical environment. Here, we analyze Illumina 1M SNP array data in three Western Pygmy populations from Cameroon and three neighboring Bantu-speaking agricultural populations with whom they have admixed. We infer genome-wide ancestry, scan for signals of positive selection, and perform targeted genetic association with measured height variation. We identify multiple regions throughout the genome that may have played a role in adaptive evolution, many of which contain loci with roles in growth hormone, insulin, and insulin-like growth factor signaling pathways, as well as immunity and neuroendocrine signaling involved in reproduction and metabolism. The most striking results are found on chromosome 3, which harbors a cluster of selection and association signals between approximately 45 and 60 Mb. This region also includes the positional candidate genes DOCK3, which is known to be associated with height variation in Europeans, and CISH, a negative regulator of cytokine signaling known to inhibit growth hormone-stimulated STAT5 signaling. Finally, pathway analysis for genes near the strongest signals of association with height indicates enrichment for loci involved in insulin and insulin-like growth factor signaling.
Published in the journal: . PLoS Genet 8(4): e32767. doi:10.1371/journal.pgen.1002641
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002641Summary
African Pygmy groups show a distinctive pattern of phenotypic variation, including short stature, which is thought to reflect past adaptation to a tropical environment. Here, we analyze Illumina 1M SNP array data in three Western Pygmy populations from Cameroon and three neighboring Bantu-speaking agricultural populations with whom they have admixed. We infer genome-wide ancestry, scan for signals of positive selection, and perform targeted genetic association with measured height variation. We identify multiple regions throughout the genome that may have played a role in adaptive evolution, many of which contain loci with roles in growth hormone, insulin, and insulin-like growth factor signaling pathways, as well as immunity and neuroendocrine signaling involved in reproduction and metabolism. The most striking results are found on chromosome 3, which harbors a cluster of selection and association signals between approximately 45 and 60 Mb. This region also includes the positional candidate genes DOCK3, which is known to be associated with height variation in Europeans, and CISH, a negative regulator of cytokine signaling known to inhibit growth hormone-stimulated STAT5 signaling. Finally, pathway analysis for genes near the strongest signals of association with height indicates enrichment for loci involved in insulin and insulin-like growth factor signaling.
Introduction
The term ‘Pygmy’ is applied to human populations whose adult males exhibit an average height of ∼150 cm or less, although thresholds between 140 and 160 cm have been employed [1]. Such groups are found all over the world including Africa, Asia, and the Americas, tend to live in tropical environments, have high levels of pathogen exposure, and practice a predominantly hunting and gathering lifestyle [2]. Studies of African mtDNA diversity suggest a time to most recent common ancestor (TMRCA) between African Pygmies and West African Bantu groups of ∼60–70 thousand years ago (kya) and between Eastern (Efe, Mbuti, Batwa, Babinga, etc.) and Western (Biaka, Baka, Bakola, Bedzan, etc.) Pygmy groups ∼10 to 27 kya [3], [4], [5], [6], [7]. Approximately 4–5 kya, Bantu speaking populations practicing slash-and-burn agriculture expanded into the forest habitats populated by the ancestors of modern Pygmy populations [8]. This secondary contact resulted in admixture, predominantly involving maternal Pygmy and paternal Bantu ancestors that remains on-going [6], as well as a cultural exchange that eliminated the Pygmy's ancestral language [9].
Short stature in human Pygmy populations is thought to be adaptive and its global distribution indicates either ancient common ancestry among tropical hunter-gatherers or extensive convergent evolution [2]. Several selective mechanisms have been proposed to explain a fitness advantage for small body size in tropical, forest-dwelling, hunter-gatherer populations. These include resistance to heat stress under humid forest conditions [2], reduced caloric requirements in a relatively food-limited environment [2], and a life-history trade-off between cessation of growth and early reproduction [1], [10]. However, the timing of the putative selective episodes and emergence of contemporary Pygmy morphology and physiology remain unclear. It is also unclear how the genetic architecture of body size in these groups may differ from that observed in other West African populations [11] and Europeans, in which ∼180 loci explaining ∼10% of the phenotypic variance have been identified [12].
To date, a variety of physiological traits related to stature have been examined in African Pygmy populations [13], [14]. These studies documented normal levels of circulating human growth hormone (HGH) but altered glucose homeostasis, insulin secretion, and free fatty acid profiles following HGH, glucose, and arginine challenges [13], [14]. These observations suggest peripheral sub-responsiveness to the effects of HGH in various tissues. Subsequent work noted a significant difference in overall growth rate in Pygmies only during puberty and suggested a reduced level of IGF-1, whose production is stimulated by HGH, is more likely responsible for short stature in adults [10], [15], [16]. In addition, both a reduction in IGF-1 receptor expression and decreased signal transmission in response to physiological concentrations of IGF-1 were observed in immortalized Pygmy T-cells [16], [17]. The observations of low levels of high-affinity HGH binding protein in both African and Philippine Pygmies [18], [19], and a severe under-expression of HGH receptor (HGHR) in the Bakola Pygmies [20] suggest a deficiency in HGH receptor activity contributes to short stature as well.
With the goal of providing a genomic perspective on the adaptive and genetic basis of short stature in Pygmies, we analyzed genetic and phenotypic data in a set of neighboring Western Pygmy (Baka, Bakola, and Bedzan) and Bantu-speaking (Tikar, Ngumba, Lemande) agricultural populations from Cameroon. Our sample includes 67 Pygmy and 58 Bantu individuals genotyped using the Illumina 1M SNP array. Of these, 57 Pygmy and 39 Bantu individuals have corresponding phenotype data for height. We performed several genome-wide scans for positive selection, including the calculation of FST and locus specific branch length (LSBL) metrics, as well as iHS and XP-EHH analyses. Since regions exhibiting selection signals are likely enriched for functional variants contributing to adaptive differences in phenotype between populations, we summarize the types of loci found in these regions using pathway enrichment analyses. Finally, using both SNP genotypes directly as well as ancestry estimates that facilitate admixture mapping approaches, we performed association tests within candidate regions identified in our selection scans as well as for SNPs near genes in candidate signaling pathways. This multifaceted approach identified several genomic regions showing strong signals of selection and association, some of which contain candidate genes for height variation. Together, our results provide unique insights into both the complex adaptive process that took place in the ancestors of contemporary Western African Pygmies and Bantu agriculturalists and the genetic architecture of short stature in Western African Pygmies.
Results
Population Structure
We observed extensive and significant genetic and phenotypic differentiation (Figure 1, Figure 2, Figure S1) and varying levels of admixture among the Pygmy and Bantu populations. Average levels of Bantu ancestry, as determined by STRUCTURE (K = 2), in the three Western Pygmy populations were 27% (Bakola), 35% (Baka), and 49% (Bedzan) with individual values ranging from 16–73%. Average levels of Pygmy ancestry in the three Bantu populations were <1% (Lemande), 2% (Tikar), and 7% (Ngumba), with individual values ranging from 0–39%. We also observed a highly significant correlation between ancestry and height (p = 5.047×10−18) after correcting for the effect of sex (full model r2 = 0.7411, r2 for sex = 0.4247; r2 for ancestry = 0.3164). In addition, the effect of ancestry remains significant in a model that also includes Pygmy-Bantu ethnicity as a covariate (p = 3.8×10−5). These results are consistent with Becker et al. [21] and indicate a strong genetic influence on height. Similar findings were also observed using Pygmy samples only (pancestry = 0.000216; full model r2 = 0.5066; r2 sex = 0.3744; r2 ancestry = 0.1322) and the independent set of genome-wide microsatellite markers described in Tishkoff et al. [9] (data not shown).
Fig. 1. Admixture and population structure. 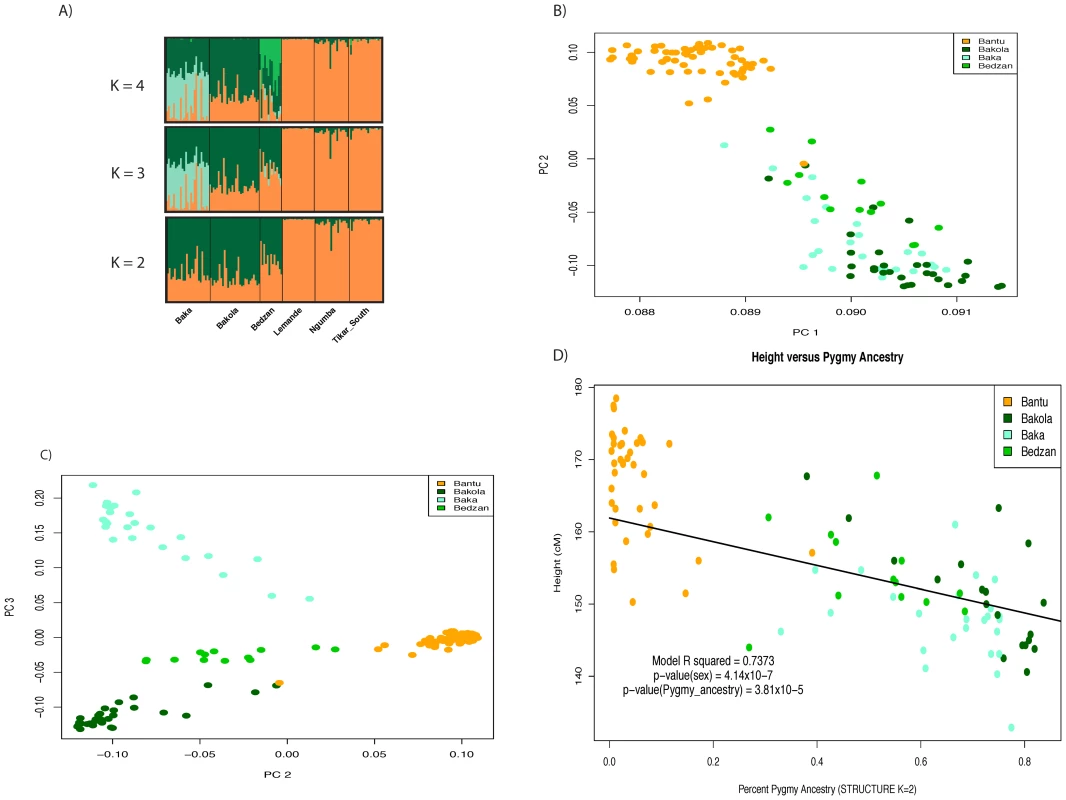
Plots of principal components A) 1 versus 2 and B) 2 versus 3 calculated using 1M SNP data from 67 Pygmy and 58 Bantu samples. The proportion of variance explained by PCs 1, 2, and 3 is 0.245, 0.0192, and 0.0109, respectively. The three Pygmy ethnic groups are clearly distinguished from their neighboring Bantu populations by the first two PCs, while the third differentiates the three Pygmy groups from one another. C) Regression of height on Pygmy ancestry inferred using STRUCTURE (K = 2). Higher levels of Pygmy ancestry are associated with shorter stature. The trend line intersects the intercept from the full model and has a slope equal to the regression coefficient for percent ancestry. D) Visualization of percent ancestry inferred using STRUCTURE at K = 2, 3, and 4. Fig. 2. Results from local ancestry inference. 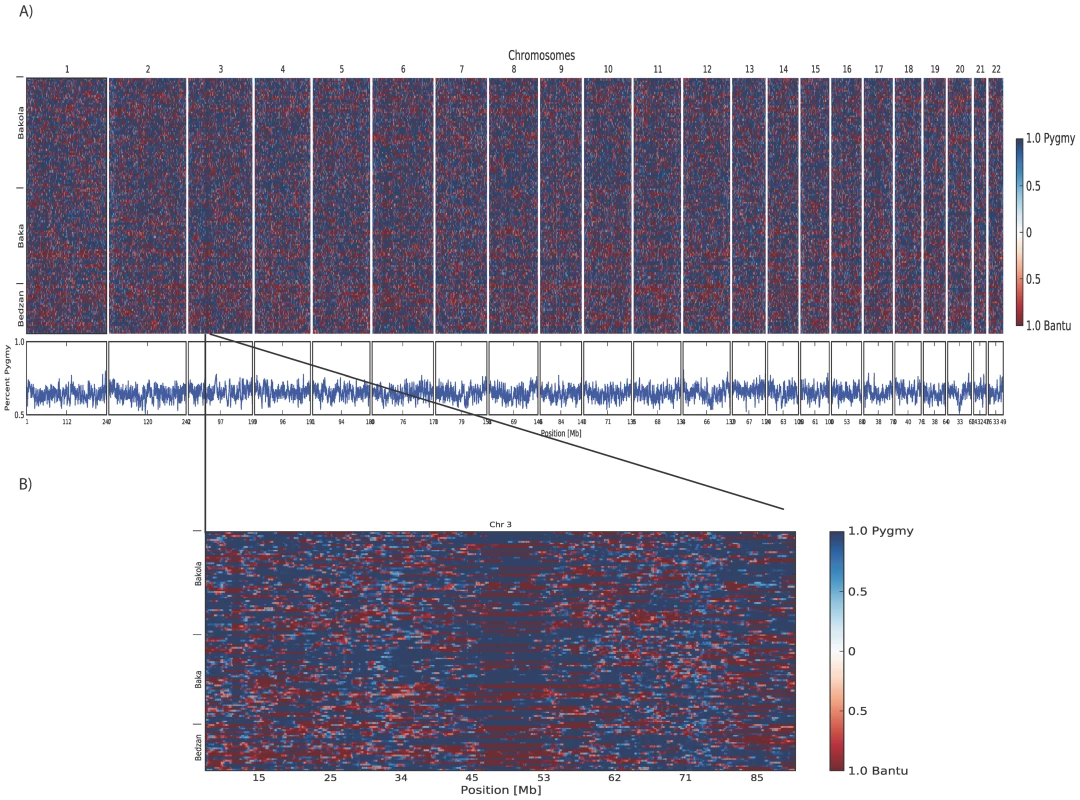
A) Genome-wide blocks of Pygmy and Bantu ancestry in Pygmy individuals. B) Blocks of Pygmy and Bantu ancestry in Pygmy individuals for chromosome 3. Shades of blue represent Pygmy ancestry and shades of red admixed Bantu ancestry. Shades are determined by the posterior probability of the estimates; darker colors indicate greater confidence. Each individual is represented by two horizontal rows (one for each haploid genome) and columns represent each window in the genome. Inferred percent Pygmy ancestry for each window is given below. Using SupportMix analysis, we determined the average ancestry block sizes in Pygmies to be 1.7+/−2.4 Mb for Pygmy ancestry, and 3.1+/−4.6 Mb for Bantu ancestry. Inference of Local Ancestry
The relatively high levels of Bantu ancestry in Western African Pygmy individuals suggests admixture mapping approaches will be useful for identifying loci underlying the short stature phenotype [22]. Thus, we inferred ancestry blocks across haploid Pygmy genomes using the machine learning method SupportMix, which does not require an a priori admixture model. Consistent with a model of ancient gene flow, we observe very short average tract lengths of Bantu ancestry (3.1+/−4.6 Mb; Figure 2A). No autosomal regions were significantly enriched for either Pygmy or Bantu ancestry (Figure 2A). However, we do observe regions with reduced levels of switching between ancestry blocks. One such region extends across a 7 Mb region from 46 Mb–53 Mb on chromosome 3 (Figure 2B).
Scans for Selection: FST/LSBL Analyses
We performed several genome-wide scans for selection that have complementary properties for detecting adaptive events on different timescales and under different selective scenarios [23]. We began by calculating pairwise FST values [24] using the Bantu (N = 44) and Pygmy samples (N = 30) estimated to have the lowest levels of admixture in our STRUCTURE analysis. Next, we identified SNPs that are differentiated specifically on the Pygmy lineage using three-way Locus Specific Branch Length analysis (LSBL [25]) comparing Western Pygmy, Bantu, and Hapmap Maasai (N = 45) populations. These cross-population scans for selection (e.g. FST and LSBL) are expected to be useful for detecting regionally restricted adaptation, including selection on standing variation [26].
Genome-wide values for pairwise FST ranged from 0 to 0.58. High FST SNPs are found on all 22 autosomes, but do not appear randomly distributed across the genome. Rather, of the 947 SNPs in the top 0.1 percentile of the empirical distribution (FST>0.33; Table S1), approximately one third (308) occur in 37 high-density (>2.31/Mb) blocks that average 93 kb in length. Such blocks are defined as a series of four or more high FST SNPs with less than 100 kb separating each and occur on all chromosomes except 11, 13, 16, 20 and 22. Using these criteria, 33% of the most differentiated loci in the genome are found in a total of just 3.42 Mb. The most striking signal (34.7 high FST SNPs/Mb) occurs within a 15 Mb region containing 9 separate blocks on chromosome 3 (between 45 and 60 Mb), 7 of which occur between 48 and 53 Mb (Figure 2B, Figure 3, Figure 4A–4E). Interestingly, the highest FST SNP in the region for which there is genotype data from the CEPH human diversity panel (rs7626978 at position 48505831; FST = 0.53 [27]) shows a striking global distribution in which the minor allele is most common in the Western and Eastern Pygmy, San, and neighboring populations and is nearly absent outside of Africa (Figure S2).
Fig. 3. Genome-wide clusters of signals of natural selection in Pygmies. 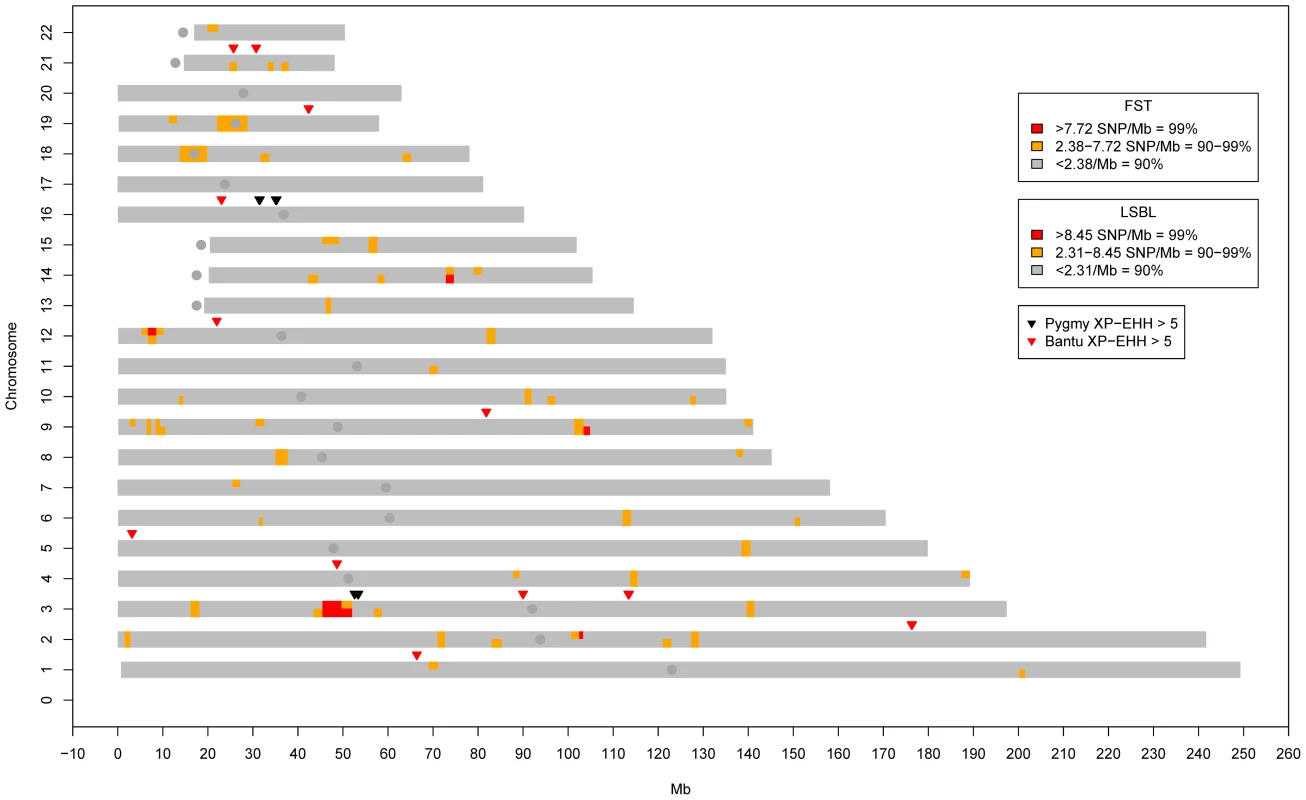
Blocks of high genome-wide density based on SNPs/Mb for FST (top half of each chromosome) and Pygmy LSBL SNPs (bottom half of each chromosome) are shown. Also shown are XP-EHH signals >5.0 in the Pygmy (Black) and Bantu (Red). The only genomic location showing all three signals occurs on chromosome 3 roughly between 45 and 60 Mb. iHS results for this region are given in Figure 4C and 4D. Fig. 4. Signals of positive selection and association with height on chromosome 3 between 45 and 60 Mb. 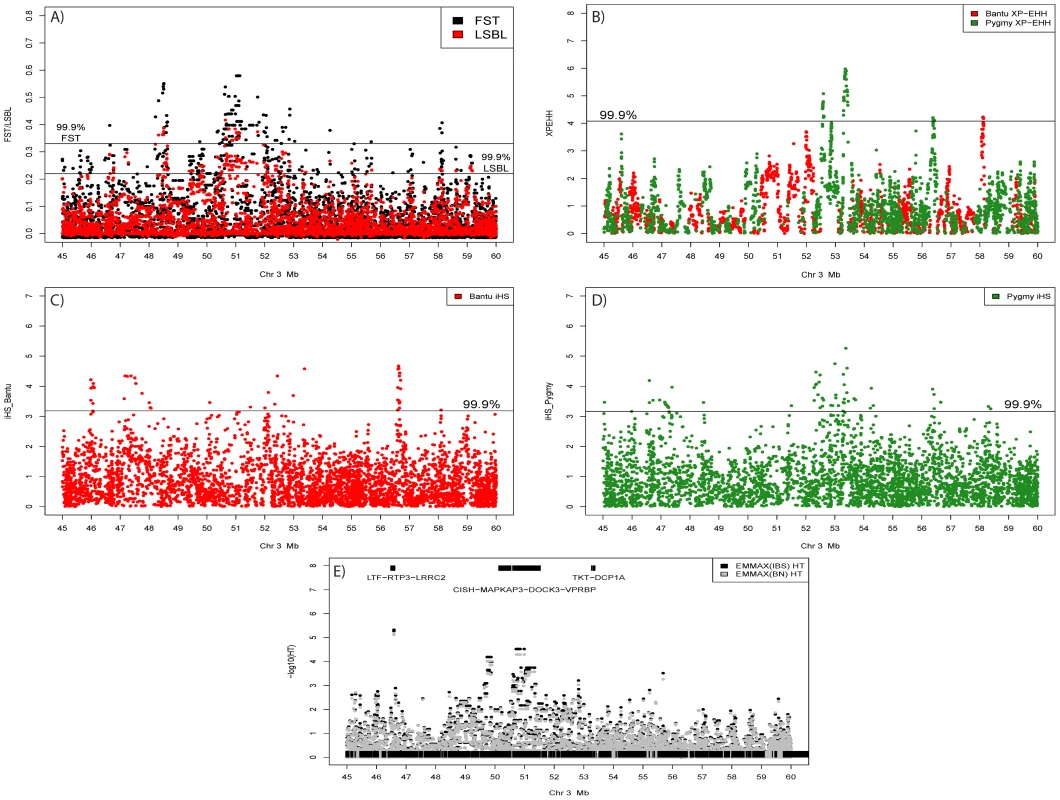
A) Results from FST (Black) and Pygmy LSBL (Red) analysis. The region between ∼50 and 52 Mb contains the largest concentration in the genome of SNPs with scores in the 0.1% tail of the empirical distribution and contains the positional candidate genes CISH, MAPKAP3, and DOCK3. B) Results from XP-EHH analysis. Scores depicted in red show negative values indicating a signal in the Bantu samples, and scores in green show positive values indicating a signal in the Pygmy samples. C) Results from iHS analysis in the Bantu samples. D) Results from iHS analysis in the Pygmy samples. E) Results of association analysis for height (cm) using EMMAX with sex as a covariate. Two suggestive associations are evident. The first, at ∼46.5 Mb, occurs within the coding region of LRRC2. The second is a strong enrichment of association signals between ∼49.5 and 51 Mb directly corresponding to Pygmy-specific signals of positive selection in A–D, centered on the three positional candidates CISH, MAPKAP3 and DOCK3. Genome-wide values for Pygmy-specific LSBL SNPs ranged from 0 to 0.42. High Pygmy LSBL SNPs representing the top 0.1 percentile of the empirical distribution (912 with Pygmy LSBL>0.2215) are found on all 22 autosomes (Table S2) and a large proportion of these (306 SNPs, ∼34%) are found in 38 physical clusters (visualized as regions of density >2.31 high Pygmy LSBL SNPs/Mb in Figure 3) averaging 99.85 kb in length. Many overlap with high FST clusters, as expected. Nine of the 38 Pygmy-specific LSBL regions also occur on chromosome 3 between 45 and 60 Mb.
In order to test the null hypothesis of a random distribution of signals over the genome, we defined non-overlapping 500 SNP bins (average size ∼1.77 Mb) and assessed the significance of the number of high FST and high Pygmy LSBL SNPs observed in each using the binomial distribution. We identify 82 and 81 bins with over-representation of high FST and high Pygmy LSBL SNPs, respectively, that are significant at the p<0.01 level. Among these are three contiguous bins (47560863–49772375 bp, 49773236–52166147 bp, and 52169095–53855083 bp) in the chromosome 3 region showing p-values less than 1×10−12 (numbers of high FST SNPs = 18, 83, and 11; numbers of high LSBL SNPs = 22, 92, 16). Together, these three bins account for 11.8% and 14.3% of all high FST and high Pygmy LSBL markers in the genome.
FST and Pygmy-specific LSBL values for X chromosome SNPs are given in Table S3A, S3B). As expected due to its smaller effective population size, these values are higher on average than values calculated for the autosomes. We identify 31 high FST and 11 Pygmy specific LSBL SNPs within the top 0.1% of X chromosome values, none of which are near obvious candidate genes for height. Because of the distinct demographic history of the X chromosome, we focus further analyses on only autosomal SNPs.
Scans for Selection: XP-EHH and iHS Analyses
Next, we performed iHS [28] and XP-EHH tests of neutrality [29] in the full set of samples to identify regions that may have been targets of recent selective sweeps within and between populations, respectively. Using an XP-EHH threshold of 4.08 (top 0.1%) we identified 400 SNPs, 54 of which fall in the 45–60 Mb region of chromosome 3. The others are spread over the remaining autosomes with the exceptions of chromosomes 15, 18 and 22. Using an even more stringent threshold of 5.0 (the top 0.02% of the distribution) we identified six Pygmy-specific XP-EHH signals, two of which occur on chromosome 3 between 45 and 60 Mb (Table S4; Figure 3, Figure 4). Using iHS we identified 784 SNPs in the Pygmy population and 807 SNPs in the Bantu population showing signals of selection that are widely distributed across the genome (Tables S5, S6). In the Pygmies, 25 of the top iHS SNPs are again found between 45 and 60 Mb on chromosome 3.
Pathway Enrichment Analysis of Scans for Positive Selection
In order to summarize the types of loci and explore the potential adaptive genetic architecture implicated by our genome-wide selection scans, we identified all protein coding genes within 100 kb up - and downstream of SNPs showing signatures of selection. Using PANTHER, we then identified pathways that appear enriched in each analysis (Table S7). Significant enrichment for neuro-endocrine signaling and pathways potentially involved in reproduction and thyroid function, including oxytocin receptor mediated signaling and thyrotropin-releasing hormone receptor (TRHR) signaling, was found for genes near high Pygmy LSBL SNPs (Table S7A). Oxytocin plays a role in lactation, parturition, and pair bonding [30] whereas the TRHR signaling pathway plays a role in regulation of the hypothalamic-pituitary–thyroid axis and influences growth, thermo-regulation, reproduction, and immune response [31]. The observation of a relatively low incidence of goiter in Eastern [32] and Western Pygmies (personal communication, B. Hewlett and L. Cordes) relative to neighboring non-Pygmy populations raises the possibility that Pygmies possess a biological adaptation to a low iodine environment [32], [33], consistent with the observation that genes involved in thyroid function are targets of selection in Pygmies [33]. Though there is little significant pathway enrichment for genes near Pygmy-specific XP-EHH and iHS signals (Table S7B–S7C) after correction for multiple tests, several suggestive enrichments are observed for pathways that play a role in immunity and neuroendocrine function. Several of these enrichment categories overlap across different analyses (e.g. 5HT2 type (serotonin) receptor mediated signaling, oxytocin receptor mediated signaling, and TRHR signaling pathways). Finally, a more formal test for enrichment of selection signals at SNPs near genes in the candidate HGH/IGF1 pathway [34] indicated a significant enrichment (p<0.0001) of iHS signals. Similar analyses for FST, LSBL, and XP-EHH were non-significant.
Signatures of Selection and Association at GWAS Significant SNPs for Height in Non-Africans
A total of 112 of the 211 SNPs significantly associated with height variation in recent GWAS in non-Africans [12] are also present on the 1M chip, passed QC, and are polymorphic in our study populations (Table S8). Interestingly, the highest FST calculated for a non-African GWAS SNP in our study was 0.23, a value far below our high FST cutoff, and most (N = 94) showed FST values<0.10. Only 5 show values greater than the approximate worldwide average of 0.15 and only 7 occur within 100 kb up - and downstream of a high FST SNP in our study populations. Similarly, the highest Pygmy-specific LSBL value for these markers was 0.14, also far below our 0.1% cutoff. In addition, the maximum absolute value XP-EHH score for a non-African GWAS significant SNP was 2.68, much lower than our cut-off of 4.08. However, one non-African GWAS significant SNP shows an iHS score in the 0.1% most extreme values in the Pygmies (rs16964211 = 3.233). The positional candidate associated with this SNP is CYP19A1, a member of the cytochrome P450 superfamily. Overall, these findings are consistent with Pickrell et al [35], which reports little evidence for selection at these loci in Western and Eastern Pygmy populations. Additionally, only four of these GWAS significant SNPs show raw p-values for association with stature less than 0.05 in our combined sample (rs1351394 = 0.01978097, rs724016 = 0.02810565, rs9969804 = 0.04384069, rs4630309 = 0.04465145).
Because of the possibility that the European GWAS associated markers and nearby functional variants identified in non-Africans are unlinked in our African populations, we also compared the positional candidate genes reported in non-African GWAS (rather than the SNPs themselves) to a list of all genes within 100 kb windows up - and downstream of signatures of selection in our study. We observe 69 candidate genes for stature that fall within such windows, including DOCK3 (33 occur near Pygmy-specific signatures of selection, Table S9). However, 50 candidate genes from GWAS were observed in 2000 randomly sampled, genome-wide 200 kb bins. Thus, observing 69 such loci in the approximately 3500 such bins identified in our analyses does not appear exceptional. These results suggest that the genetic architecture of height in Pygmies differs from that observed in Europeans; however some overlap in specific genes involved remains a likely possibility.
Admixture Mapping in West African Pygmies
In order to incorporate ancestry information directly into association testing, we performed admixture mapping [22] using EMMAX. Specifically, we tested high confidence ancestry blocks inferred in our Pygmy samples (N = 57) by SupportMix (unpublished data) for association with height variation. This analysis identified several suggestive associations with stature (p<0.002; Table S10), though none show striking FDR-corrected p-values. The strongest (p = 1.40×10−5; FDR = 0.304) is observed for a 50 SNP ancestry bin on chromosome 2 approximately 268 kb from the gene APOB. Interestingly, studies have demonstrated a correlation between high APOB levels and short stature [36].
Targeted SNP–Based Association Analysis for Height in Western African Pygmies
In an approach adapted from Moore et al. [37], we next performed targeted association tests, first in Pygmy individuals only, for SNPs showing Pygmy-specific signatures of selection (extreme high Pygmy LSBL, Pygmy XP-EHH and Pygmy iHS; N = 2288 SNPs). These marker-based association analyses identified several suggestive associations with height (Table S11A), though they were not significant after multiple tests correction. The marker showing the strongest association (p = 0.0004; FDR = 0.241) is found in a cluster of immunoglobulin lambda loci. The second strongest association (p = 0.0005; FDR = 0.241) occurs near the gene NDUFA4. While a direct connection to height variation is not obvious for NDUFA4, it is a component of the mitochondrial respiratory chain, which plays a role in metabolic function. The next several strongest associations are found in or near the gene MAP3K2, which is involved in NF-kappa B signaling and the regulation of JNK and ERK5 pathways which play a role in growth factor signaling. Additionally, five of the strongest association signals are observed for markers within the 49–53 Mb region of chromosome 3 (Table S11A).
Since growth hormone and IGF-1 signaling are strongly implicated in Pygmy-specific phenotypic variation by extensive physiological studies in African Pygmies, we next tested SNPs 100 kb up - and downstream of genes in HGH, IGF-1, and INS signaling pathways (N = 40,558) for association with height variation in the Pygmy population. The markers showing the strongest associations with height near GH-IGF1-INS genes are in or near the genes GCK (p = 6.08×10−5; FDR = 0.591), DUSP4 (p = 7.82×10−5; FDR = 0.591), and IGFALS (1.20×10−4; FDR = 0.591), although none of these associations are significant after correction for multiple tests (FDR<0.05).
Targeted Association with Height Variation in the Combined Pygmy–Bantu Sample
We next performed association tests in the larger combined Pygmy-Bantu dataset for SNPs showing Pygmy-specific signatures of selection. This both maximizes our sample size and takes advantage of the extensive between-population phenotypic variation. We observed 128 SNPs (Table S11B) showing significant associations after correction for multiple tests (FDR<0.05) and for population substructure. The strongest significant associations are found in or near the genes NDUFA4 (p = 1.31×10−5; FDR = 0.007), ATP2B4 (p = 2.73×10−5; FDR = 0.007), and DOCK3 (p = 2.99×10−5; FDR = 0.007) (Table S11B, Figure 4E). Interestingly, NDUFA4 is also implicated in the Pygmy-only analysis above. The strongest association signal in DOCK3 consists of 8 linked SNPs spread over ∼268 kb that are found in the middle of the genomic region on chromosome 3 (45 Mb–60 Mb) displaying multiple signals of positive selection (Figure 4A–4D) and association in the Pygmy population (Table S11A). They also occur ∼85 kb from a marker (rs13088462) showing significant association with height variation in Europeans [12]. Two additional SNPs flanking this European GWAS-associated SNP by ∼4 kb on either side (rs4443210 and rs7638732) also show significant association with height variation in our analysis (p = 0.00018 and 0.00023; FDR = 0.013 and 0.013, respectively).
We next tested SNPs 100 kb up - and downstream of genes in HGH, IGF-1, and INS signaling pathways for association with height variation in our combined Pygmy-Bantu samples (Table S12). The strongest associations are found in or near CISH (p = 2.99×10−5; FDR = 0.199), LEPR (p = 3.36×10−5; FDR = 0.199), EEF2K (p = 3.44×10−5; FDR = 0.199), PRL (p = 6.38×10−5; FDR = 0.323), and IFNG (p = 8.80×10−5; FDR = 0.397), though none were significant after FDR correction for multiple tests.
Pathway Enrichment Analysis in Extreme Genome-Wide Association Signals
Though our small sample size limits power to detect significant associations using full genome-wide association analyses (see Figure S3), we explored potential pathway enrichment for genes 100 kb up - and downstream of markers in the extreme tail of the empirical distribution of genome-wide p-values (the lowest 0.1%) (Table S7D). Consistent with the physiological literature in African Pygmies, the top two pathways that are significantly enriched in the combined Pygmy-Bantu association analysis are the protein kinase B signaling and the mitogen activated protein kinase/MAP kinase cascades of the Insulin/IGF pathway (Table S7D). While PANTHER enrichment for these pathways does not reach statistical significance after Bonferroni correction for multiple tests, the likelihood that these two pathways are the two most enriched by chance is very low, as evaluated using a non-parametric re-sampling test (p = 2×10−4).
Discussion
Our analyses suggest that a complex set of demographic and selective dynamics have shaped genetic and phenotypic variation in West African Pygmies and neighboring agricultural groups. For example, the very short average tract lengths of inferred ancestry we observe (average Bantu tract length of 3.1+/−4.6 Mb) are strikingly different from those seen in simpler admixture scenarios (e.g. African Americans). The shorter average Bantu tract length we observe appears to reflect the long history of admixture between Western Pygmy and neighboring Bantu populations that has taken place, possibly since the Bantu expansion into the rain forest several thousand years ago ([8]). Although models assuming a scenario of a single recent pulse of admixture have been developed for inferring the time of admixture based on the distribution of admixture tract lengths [38], more complex models will need to be developed to make inferences about ancient admixture events (e.g. >100 generations) which may fluctuate in rate over time [39].
While the increased genetic resolution offered by short tract lengths may eventually prove useful in fine mapping the genetic basis of complex traits, it also complicates the application of standard approaches to ancestry inference. The novel, model-free SVM approach to ancestry inference we apply here performs well compared to standard approaches in admixed Western Pygmy populations. However further refinement of ancestry inference methods designed to deal with complex admixture histories as well as multiple and/or unknown ancestral populations are needed. Additionally, the development of unbiased African SNP genotyping arrays (e.g. those not based on SNPs and patterns of LD identified in non-African populations) are likely to provide novel insights into genome-wide patterns of ancestry and, coupled with increased samples sizes, will facilitate association studies of complex trait variation in African samples.
Signatures of Selection
Our multiple tests of neutrality, which are sensitive to different selection scenarios and timescales, revealed many signatures of positive selection which are enriched for genes influencing a variety of complex traits with potential fitness implications including body size, thyroid function, immune system function, and reproduction. Although we see evidence for enrichment in HGH pathway genes in our Pygmy-specific iHS results, suggesting some selection on these loci after the split between Eastern and Western pygmies (>10 kya), the genome-wide distribution of signals is not consistent with a strong, recent selective sweep at a single locus. Rather, the overall results of our selection scans suggest that the process of local adaptation involved multiple loci that may have been favored at different points in time. Additionally, targets of selection from standing variation or those showing slight changes in allele frequency at multiple loci are challenging to identify. Further development of methods sensitive to these types of complex adaptive signatures will greatly facilitate the analysis of the genetic architecture of adaptation in these and other global populations [40].
The Chromosome 3 : 45–60 Mb Region
The region found on chromosome 3 between 45 and 60 Mb shows an overlapping pattern of selection and association signals (Figure 4). The presence of multiple, Pygmy-specific signals of selection using different tests of neutrality could indicate that the region has been repeatedly under selection during the process of local adaptation in Western Pygmy populations. However, the additional presence of Bantu-specific signatures of selection suggests that the region may be more broadly relevant for adaptation in human populations. Indeed, this region has been previously implicated in genome-wide scans for selection in Hapmap populations from Europe, Asia, and Africa [23], [28], [29], [41], [42], [43]. SNPs in this region also show significant association with height variation in both our combined Pygmy-Bantu samples and in recent GWAS studies in Europeans as well as suggestive association in Pygmy samples only (Tables S11 and S12).
Among the most promising positional candidates in the chromosome 3 region are DOCK3, a guanine nucleotide exchange factor shown to be associated with height variation in non-African populations [44], MAPKAP3 a mitogen-activated protein kinase central to several signaling pathways [45], and CISH, a negative regulator of cytokine signaling that also inhibits HGHR activity [46]. The 709 kb long DOCK3 gene is expressed specifically in the brain [47] and has no obvious impact on stature, although a SNP within DOCK3 is associated with height in Europeans [12]. However, ∼63 kb away from DOCK3, oriented in the opposite direction, is the 5.4 kb CISH gene. CISH, a member of the cytokine signaling (SOCS) family of proteins, is up-regulated by interleukin-2 (Il-2) and plays a critical role in the signaling of cytokines by binding tyrosine residues on activated cytokine receptors, particularly IL-2R [48], as well as T-cell differentiation [49]. A recent study demonstrated that genetic variation at CISH is associated with susceptibility to bacteremia, malaria, and tuberculosis in several global groups including a Gambian population [48], indicating the important role that CISH plays in infectious disease susceptibility. CISH also directly inhibits HGHR action by blocking the STAT5 phosphorylation pathway [49], and CISH expression is highly regulated by levels of HGH expression [50]. Transgenic mice that over-express CISH show reduced growth and overall small body size [49].
Sequencing of 5 kb of the CISH gene inclusive of the protein coding region, (Figure S4A–S4C) reveals increased diversity in Pygmy individuals relative to the Bantu individuals, but no deviations from neutral expectations. We also did not identify any common population-specific SNPs. However, it is possible that a genetic variant upstream of CISH, possibly within or near the DOCK3 gene, contains a regulatory element that alters CISH gene expression in Pygmies.
Although we did not detect any structural variants (SVs) in the region using SNP data (data not shown), DOCK3 is known to contain inversion polymorphisms in global populations between ∼50.88–50.97 Mb [51]. This observation raises the possibility that an undetected SV could play a role in altering gene expression in the region. Indeed, 8 SNPs among the most strongly associated with stature in our combined Pygmy-Bantu analysis occur in the DOCK3 gene and are in 100% LD over a ∼268 kb region in both Pygmy and Bantu populations. Specifically, after phasing, genotypes at these SNPs are found as two “scaffold” haplotypes: AAGGGAAG and GGAAACGA, with the “A” form (those with an A at the first SNP, rs6779819) at ∼73% frequency in the Pygmy sample, and ∼36% in the Bantu. These markers also show strong linkage disequilibrium in HapMap European and Asian samples where the same “A” form haplotype is found at 26% and 38% frequency, respectively. Additionally in our Pygmy-Bantu analyses, variation at intervening marker loci define lower-frequency haplotypes, several of which are private (or near private) in the Pygmy population across the region (right half of Figure S5B). This pattern of variation is consistent with the possibility of an ancient inversion encompassing the 8 “scaffold” SNPs that has restricted recombination in the region, with the subsequent accumulation of variation resulting in each sub-type. Moreover, we observe multiple long-range population-specific haplotypes (∼1 Mb in size; Figure S5) in the CISH/MAPKAP3/DOCK3 region in both the Bantu and Pygmy populations that likely contribute to the high FST signal (Figure 4A). Additionally, we observed reduced levels of switching between ancestry blocks in the 46 Mb–53 Mb region on chromosome 3 (Figure 2B), consistent with reduced recombination. Future functional studies will be required to determine the magnitude of any CISH expression differences between Pygmies and other populations and how these might alter the downstream effects of HGH in African Pygmies.
In addition to CISH, MAPKAP3, and DOCK3 there are several other positional candidate genes in the chromosome 3 region between 45 and 60 Mb that may have played a role in Western Pygmy adaptation. These include ERC2, at position ∼55 Mb, which occurs within 100 kb of signals in all five scans for selection (Table S9). Together with BSN located at ∼49 Mb in the chromosome 3 region, ERC2 appears to help regulate neurotransmitter release [52]. This raises the interesting possibility that an interaction between these loci, possibly related to reproduction, is contributing to the strong Pygmy-specific differentiation in the region. Thus, it is possible that a co-adapted complex of genes in the chromosome 3 : 45–60 Mb region has been under selection in the Western Pygmy lineage, as has been observed in other species in regions with reduced recombination due to structural variation [53].
Pathway Enrichment
The pathway enrichment results from our selection scans and height association results provide suggestive, novel insights into the possible genetic and physiological architecture of adaptation in Western African Pygmies. Signatures of selection in Western Pygmies are often found near genes involved in neuro-endocrine signaling pathways related to reproduction and steroid hormone synthesis, supporting the proposal that differences in the timing of reproduction may partly explain short stature as adults in these populations [1]. Further study of specific genes in these pathways and their expression patterns are likely to yield novel insights about their functional role in adaptation in Pygmies. In addition, the enrichment for genes that play a role in the Insulin/IGF pathway near the strongest signals of genome-wide association further supports a major role for this pathway in the genetic architecture of height in Western African Pygmies.
Conclusions and Future Studies
Our multi-dimensional approach incorporating ancestry estimation, genome-wide scans for positive selection, and genotype/phenotype association identified several candidate genes and pathways that may contribute to adaptive phenotypes, including short stature, in Western African Pygmy populations. Our results raise the possibility that the adaptive process that produced small body size in Pygmies may be the result of selection for traits other than stature, including early reproduction, metabolism, and immunity, and that the functional variants (possibly regulatory in nature) that are targets of selection may have pleiotropic effects. Indeed, given the extremely high pathogen load and short life span in Pygmies (between 15.6 and 24.3 years [1]) immune function is likely to have been under strong selective pressure in Pygmy populations. Furthermore, many of the loci that play a role in immune response (e.g. cytokines) are known to directly influence genes related to HGH and insulin/insulin-like growth factor signaling [54]. It is also possible that there have been multiple selective events in the history of the Pygmy populations, at different time periods, that may have contributed to the co-adaptive evolution of loci that play a role in immunity, metabolism, and neuro-endocrine function. Comparison of selection and association signals in Western and Eastern Pygmies will be informative for distinguishing if selection for short stature evolved in an ancestral Pygmy population prior to their divergence within the past ∼25,000 years [3], [4], [5], [6], or whether this trait evolved independently in these populations due to convergent evolution. In addition, future analysis of whole genome sequence, characterization of structural variants, and functional studies of gene expression after challenge with HGH and IGF-1 in appropriate cell types will be particularly informative for identifying genomic variants that play a role in short stature in Pygmy populations.
Materials and Methods
Subjects and Samples
Appropriate IRB approval for this project was obtained from both the University of Maryland and the University of Pennsylvania. Prior to sample collection, informed consent was obtained from all research participants, and permits were received from the Ministry of Health and National Committee of Ethics in Cameroon. In total, our study included 132 individuals from six populations residing in Cameroon. Three are Niger-Kordofanian Bantu-speaking hunting and gathering populations: the Baka, Bakola, and Bedzan, and three are neighboring Niger-Kordofanian Bantu-speaking agricultural populations: the Ngumba, Southern Tikar, and Lemande. While the term ‘Pygmy’ has historically been pejorative, more recently it has been used by indigenous groups themselves as well as activist groups working on their behalf [55], [56], [57]. In acknowledgement of this recent trend and in the absence of a better term that encompasses the hunting and gathering peoples from Cameroon, we use ‘Pygmy’ to collectively refer to the set of three populations, the Baka, the Bakola, and the Bedzan, included in our study and their ancestors. Likewise, we will use the term ‘Bantu’ to refer to the three agricultural populations in our study, the Lemande, the Ngumba, and the South Tikar, and their ancestors. Samples were collected in villages in the Eastern Province (Baka and neighboring Bantu groups), Southern and Ocean Provinces (Bakola and neighboring Bantu groups), and Center Provinces (Bedzan and neighboring Bantu population). White cells were isolated in the field from whole blood with a salting out procedure modified from [58] and DNA was extracted in the lab with a Purgene DNA extraction kit (Gentra Systems Inc., Minneapolis, MN). Height measurements as well as self-identified ethnicity, parent and grandparent information were also recorded except in the case of the Lemande for which no phenotypic measurements are available.
Genotyping and Quality Control
DNA samples were genotyped on the Illumina Human 1M-Duo DNA analysis BeadChip. All DNA samples showed SNP call rates of at least 95%. In order to ensure the quality of genotype calls, SNPs with more than 5% missing data were excluded from analysis. The remaining autosomal dataset includes a total of 1,083,730 SNPs of which 960,497 were polymorphic in our populations of interest. An additional 40,949 X chromosome SNPs exceeded similar quality control metrics and were analyzed separately to determine FST and Pygmy LSBL values. In order to identify close genetic relatives in the autosomal dataset, we estimated identity by descent (IBD) using PLINK v 1.07 [59]. Seven individuals (4 Bakola, 2 Bedzan, and 1 Tikar South) were closely related to one or more individuals in the sample. After their removal no pairwise estimate of pi-hat exceeded 0.25, a value roughly corresponding to relationships closer than grandparent-grandchild. The remaining set of 125 samples (67 Pygmies and 58 Bantu) served as the basis for subsequent analyses.
Estimates of Population Structure and Ancestry
In order to evaluate population structure in our sample, we identified a subset of SNPs that maximizes the number of markers while minimizing linkage disequilibrium (LD). Specifically, we removed SNPs within a sliding window (100 SNPs per window and an offset of 50 SNPs) based on the variance inflation factor [59] (VIF = 2). The resulting 30,404 markers were analyzed using STRUCTURE to estimate the proportion of individual ancestry for a range of ancestral clusters, K = 2, 3, and 4 (Figure 1A). Principal components analysis (PCA) was performed using R (Figure 1B, 1C; R Development Core Team [60]).
Correlation between Global Ancestry and Height Variation
We explored the relationship between global admixture and height variation using linear regression. Estimates of ancestry were analyzed using the following model:Where G is a co-variate representing sex and P represents the proportion of Pygmy ancestry. Age was initially included but was not significant and so was removed from the final analysis (Figure 1D).
Pairwise FST and LSBL analyses
First, using the subset of least-admixed Pygmy (N = 28) and Bantu (N = 40; Baka = 17, Bakola = 13) individuals identified in our STRUCTURE analysis, we calculated pairwise FST [24] values for all SNPs that passed quality control. Cross-population scans for selection (e.g. FST and LSBL) are expected to be useful for detecting regionally restricted adaptation, including selection on standing variation [26]. SNPs with FST values greater than 0.3308 represent the 99.9% tail of the empirical distribution and are referred to as “high FST SNPs”. Next, in order to identify allele frequency differences specific to the Pygmy lineage, we incorporated data from unrelated individuals from the HapMap Phase 3 Maasai sample (N = 45) [61]. We then calculated Pygmy locus-specific branch length (LSBL) values [25] for each SNP that appeared in both datasets. Values greater than 0.2215 represent the 99.9% tail of the empirical distribution and are referred to as “high Pygmy LSBL SNPs”. In order to better visualize both the clustering and concurrence of FST/LSBL signals, we calculated and plotted the density of high FST and high Pygmy LSBL SNPs per Mb for all non-overlapping 500 SNP windows across the genome (Figure 3). A matrix of average pairwise FST values between each population is given in Table S13. We were also interested to see if any unusual patterns of population structure exist on the X chromosome, and so analyzed these data separately. The Illumina software package genome studio (Illumina, Inc, San Diego, CA) was used to call SNPs in the female individuals only, and once this was accomplished, males were re-incorporated to establish hemizygous allele calls.
XP-EHH and iHS Analyses
Using binary executables made publically available by the Pritchard laboratory at the University of Chicago, we performed both cross-population extended haplotype homozygosity (XP-EHH) [29] and integrated haplotype score (iHS) [28] tests of neutrality. The XP-EHH test is most powerful when the selected allele and its associated haplotype are nearly fixed in one population but remain polymorphic or absent in a comparison population. In contrast, iHS is most powerful when a selected haplotype has reached intermediate frequency and is thus useful for detecting partial selective sweeps within a single population [29]. The iHS test has also been shown to be more robust than alternative methods under complex demographic histories and variable recombination rates [62].
SNP genotypes were computationally phased using fastPHASE v 1.4 [63] treating each chromosome independently. Haplotypes that minimized the fastPHASE switch error were used in subsequent analyses, as is recommended for data sets with many markers [63]. A fine-scale recombination map relevant to the African populations included in our study was generated using LDhat version 2.1 [64] and a dataset including a total of 100 unrelated samples. Specifically, we analyzed 25 males and 25 females, from each of two populations in HapMap3 Release 2 [65]: the Yoruba from Ibadan, Nigeria (YRI) and the Luhya from Webuye, Kenya (LWK). HapMap3 Release 2 (January 2009) data were used rather than the more recent HapMap3 Release 3 (May 2010), since these were phased using trios.
LDhat uses a Monte Carlo approach to sample from the posterior distribution of the population genetic parameter 4Ner for intervals between consecutive markers, where Ne is the effective population size and r is the per-generation recombination rate. The LDhat Markov chain was run for 10,000,000 iterations; 1,000,000 iterations were used as burn-in and only every 5,000th sample was retained to reduce auto-correlation among the posterior samples. This was done for both populations separately. The median of the retained samples was calculated in order to obtain point estimates for the population-specific 4Ner values. To avoid any population-specific demographic or selection effects in the recombination map, the YRI and LWK estimates were averaged. Using these population-averaged estimates, we computed genetic positions for each SNP in our data in units of 4Ner that were subsequently used in the XP-EHH and iHS tests. For SNPs present in our data but not in HapMap3, genetic distance was assumed to scale linearly within intervals.
iHS analysis requires ancestral state information. To establish ancestral and derived alleles for all SNPs in our data, we used the human, chimpanzee, orangutan, and rhesus macaque reference alleles in dbSNP131 [66]. The data were downloaded from table snp131OrthoPt2Pa2Rm2 of the University of California at Santa Cruz genome browser [67]. An allele was determined to be ancestral to the human population if the human reference matched 1) the corresponding chimpanzee allele, or 2) the orangutan allele if the chimpanzee allele was missing, or 3) the rhesus macaque allele if both the chimpanzee and orangutan data were missing. Approximately 5% of the SNPs in our data could not be assigned an ancestral state, due mainly to missing data in dbSNP131; this was consistent across all autosomes (data not shown). SNPs with minor allele frequencies less than 5% in either the Pygmy or Bantu populations were removed from the phased dataset in agreement with recent publications [e.g. 28]. SNPs with ambiguous ancestral states were also removed from the iHS analysis. A single XP-EHH process comparing the Pygmy sample to the Bantu samples was executed and two independent iHS processes, one for the Pygmy and one for the Bantu samples, were performed. The non-standardized scores returned by the XP-EHH and iHS binary executables were adjusted such that all scores had zero means and unit variances, either with respect to the entire data set (for XP-EHH, as described in reference [29]) or to SNPs with similar derived allele frequencies (for iHS, as described in [28]). The threshold values established from the 99.9% tail of each empirical distribution were as follows: absolute value of XP-EHH = 4.08, iHS_Pygmy = 3.18, and iHS_Bantu = 3.16. This imposes cutoffs that are either comparable or more conservative compared to the values used in previous studies using these tests (e.g. [29]; [28]). It is important to note that genomic regions identified using “outlier” approaches are not all expected to be targets of selection. Rather, SNPs within the tails of the empirical distribution are expected to be enriched for linkage with adaptive variants or can be false positives caused by historical demographic events [29]; [28]. We focus primarily on Pygmy-specific signals as these are of greatest interest with respect to identifying candidate loci that play a role in short stature in that population.
Local Ancestry Inference and Admixture Mapping
Common methods for admixture mapping assume recent admixture, relatively long tracts of LD throughout the genome, and the presence of both reference parental population samples [68]. While methods such as STRUCTURE are able to estimate admixture proportions well globally, they perform poorly with respect to the local ancestry assignments critical to accurate admixture mapping analysis [69]. Other methods such as LAMP [69] do significantly better in this respect, but require knowledge of allele frequencies from both ancestral populations for optimal performance [70]. HAPMIX [71] and PCAdmix [72] have similar requirements. In addition, with the exception of HAPMIX, most methods require unlinked markers. The demographic history of Western African Pygmy populations creates a unique challenge for these methods since admixture is both ancient and on-going. Thus, we expect a wide distribution of haplotype sizes including many that are relatively short. Furthermore, since every Pygmy individual in our sample shows some degree of Bantu admixture, only one of the two ancestral populations is still available and sampled (Bantu). We therefore have applied a novel method of local ancestry inference, SupportMix, that can identify relatively small tract lengths to estimate genome-wide ancestry (unpublished data). Simulations demonstrate this method is more accurate than LAMP for several different degrees of population structure (unpublished data). When LAMP-ANC was performed using information from both ancestral populations and compared to SupportMix using only a single ancestral source population, SupportMix was again more accurate (Figure S6). Thus, SupportMix is an appropriate method to use when ancestral allele frequencies are known for only one ancestral population, as is the case for the Western Pygmies.
Briefly, SupportMix is a two-step process that classifies the ancestral origin of small regions of the genome using a support vector machine (SVM) trained on the ancestral population(s). This is followed by a classification of ancestry with the aid of a Markov model. Specifically, the SVM learns the parameters of the Markov-process and the most probable ancestral state is solved by the forward-backward algorithm over the observed states. As noted above, the complex pattern of admixture observed in our Pygmy samples hinders this training step. To overcome this, an additional SVM was trained at each window on all Pygmy and Bantu samples. Those Pygmy samples most different from the Bantu as measured by the L2 norm from the dividing SVM hyper-plane, were chosen as region-specific “ancestral” Pygmies. This allowed a new support vector machine to be trained to segregate these region-specific “ancestral” individuals from the Bantu. This was then used to classify the remaining Pygmy samples as either ancestrally Bantu or Pygmy for a given genomic region. The proportion of region-specific “ancestral” Pygmies, chosen in each window was the same for every window and was chosen to obtain a global ancestry proportion matching that of STRUCTURE. This agreement is seen in Figure S7A. A window size of 50 consecutive SNPs was sufficient to separate the “ancestral” Pygmies from the Bantu leading to a high posterior probability (69% of windows above 0.9) from the Markov Process. Only these “high confidence” regions of inferred local ancestry were used in subsequent admixture mapping analyses.
Association Analyses
Association analyses using this dataset are complicated by the inclusion of individuals from two differentiated populations, the complex patterns of admixture within the Pygmies, and our small sample size. These factors greatly reduce the power of a genome-wide association approach (Figure S3). Instead, we used a combination of ancestry-based and marker-based approaches that are expected to produce non-overlapping but complementary information [73] within and between populations. We correct for multiple tests using an analysis-specific false discovery rate (FDR) approach [74], [75] and interpret the most extreme association signals as suggestive. Specifically, FDR values were calculated from the vector of p-values from each association analysis separately. First, we performed admixture mapping using high confidence ancestry blocks inferred by SupportMix in our Pygmy samples (N = 57). Second, in an approach adapted from Moore et al. [37] we performed association tests for two subsets of SNPs for which there are a priori expectations of association in Pygmy samples separately. These are 1) SNPs showing Pygmy-specific signatures of selection (extreme high Pygmy LSBL, Pygmy XP-EHH and Pygmy iHS; N = 1823 SNPs) which are expected to be enriched for SNPs in linkage disequilibrium with functional variants that play a role in recent adaptation in Pygmies and 2) SNPs 100 kb up - and downstream of genes in HGH, IGF-1, and INS signaling pathways (N = 40,558) which are strongly implicated in Pygmy-specific phenotypic variation by the extensive physiological studies in African Pygmies described above. Genes with roles in GH, IGF-1 and INS signaling were defined via STRING using the criteria: highest confidence, no more than 50 interactors, 200 additional nodes. Third, we performed similar marker-based analyses using all Bantu and Pygmy samples combined in order to maximize both phenotypic and genetic differentiation as well as sample size (N = 125). Finally exploratory genome-wide association analyses were performed using Pygmy samples only, Bantu samples only and the combined dataset (Figure S3).
In all cases, genotype/phenotype association analysis was conducted using EMMAX [76], a mixed-model linear regression approach that corrects for population structure even under cases of extreme differentiation [77], [78]. This method has been successfully used for association mapping in a sample of inbred, and highly sub-structured, dog breeds [77]. Simulations also demonstrate that EMMAX performs extremely well in correcting for population substructure [78]. The additional advantages of this mixed-model approach include simultaneous correction for both relatedness within populations and structure between them via a pair-wise matrix of genetic relationships among individuals. We obtained results using both matrices included in the current distribution of EMMAX (BN and IBS) for both SNP-based and admixture-based analyses. Since they were very similar, we focus on results obtained using the IBS matrix for simplicity. In all analyses, but especially the combined Bantu and Pygmy analysis, EMMAX appears to have provided a reasonable correction for population structure as judged by the resultant Quantile-Quantile plots and the generally low correspondence between high LOD scores and high FST (Figure S8). Further, by overlaying both a priori biological information as well as results from several tests of neutrality our approach, in effect, examines the joint distribution of several test statistics, a scenario under which power is expected to be greatly enhanced.
Pathway Enrichment Analyses
Several gene lists were generated based on the analyses above and PANTHER was used to summarize and explore types of loci and pathways that are enriched in the tails of the empirical distribution for signals of selection and association. Specifically, genes residing 100 kb up - and downstream of each high FST and high Pygmy LSBL SNP, as well as those with XP-EHH and iHS scores in the 99.9% of the empirical distribution were identified (100 kb flanking windows were chosen since targets of selection could include regulatory regions). Each was submitted to the PANTHER web tool separately (Table S7A–S7D). We also performed similar enrichment analysis for the lowest 0.1% of the empirical distribution of genome-wide p-values from marker-based association analyses (p<0.001; Table S7A) as well as all p-values<0.05 using ancestry estimates from SupportMix (Table S7B). Pathways passing the significance threshold for analysis-specific Bonferroni corrections for multiple tests are given in bold. In addition, we calculated the probability that the two Insulin/IGF signaling pathways would be the two most enriched in the genes 100 kb up - and downstream of markers in the extreme tail of the empirical distribution of genome-wide p-values (the lowest 0.1%) for association (Table S7D). Specifically, we sampled random sets of genes with HUGO symbols (N = 1248, corresponding to number of genes in the empirical data); only genes with UNIPROT identifiers were retained. UNIPROT identifiers were mapped to all pathways using the Panther database (ftp://ftp.pantherdb.org). We then calculated chi-squared statistics for each pathway in each random list and recorded which showed the two most extreme test statistics. We repeated this procedure 10,000 times and identified either the Insulin/IGF pathway-protein kinase B signaling cascade or the Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade as one of the two most enriched only twice (p = 2×10−4).
In order to more directly quantify the potential enrichment of selection and association signals in HGH pathway genes, we used an approach described by Eleftherohorinou et al [34]. Specifically we generated a cumulative test statistic, CAtest ∑s ∈HGH βg, where s is the index of all SNPs in the HGH pathway and β is the test-statistic (score) estimated for each SNP in a given analysis. If there were an enrichment of signals in the HGH pathway, we would expect CAtest to significantly deviate from 0. To assess this, we used a genome-wide re-sampling approach wherein the same number of SNPs found in the HGH pathway was randomly sampled with replacement from our set of genome-wide SNPs. We then calculated a new cumulative statistic CArand. This procedure was repeated 10,000 times to create a null distribution for each analysis. A skew-normal distribution was fit to the null using maximum likelihood, as described in Eleftherohorinou et al. [34]. A p-value for the observed statistic was calculated using the best-fit null skew-normal distribution.
CISH Re-Sequencing
We re-sequenced a 5.4 kb region encompassing the CISH gene and its promoter in 19 Bantu (6 Lemande, 7 Tikar South, and 6 Ngumba) and 23 Pygmy (9 Baka, 8 Bakola, and 6 Bedzan). We first amplified the entire region using the two primer sets: forward_1 5′-CCTAGAGGGTCACCTATAACCTACAC-3′, reverse_1 5′-CTTGCTGCTTATCCTCGTCCTTAC-3′ and forward_2 5′-CCTCTGAGAGACACTCCTATCCAT-3′, reverse_2 5′-CCTGCTGTCTATCCTCGTCCTTAC-3′ (Figure S4A, S4B). Individual PCR amplifications were visualized with electrophoresis in a 1% agarose gel and sequenced with the Big Dye Terminator cycle sequence kit (Applied Biosystems, Foster City, CA) and run on an ABI prism 3730XL (Applied Biosystem, Foster City, CA,). For primers used for sequencing see the legend of Figure S4. We performed additional sequencing of a 1.2 kb region to evaluate the reported functional single nucleotide polymorphisms −639, −292 and −163, described by [48] using the primers: forward 5′-GTCCGCATAACGGGAGCAACAC -3′ and reverse 5′-CGCTTACCCCTGAACGCAGAGGACC -3′. CISH sequence results were analyzed and evaluated with the software package Sequencher (http://www.genecodes.com/). Haplotypes were estimated with the software package Phase [79], [80], [81]. Network analysis was performed using the program Network (http://www.fluxus-engineering.com) using the median joining methodology (Figure S4C) [82]. DNAsp (http://www.ub.edu/dnasp/) was used for calculating nucleotide diversity and Tajima's D (Figure S4D) [83].
Supporting Information
Zdroje
1. MiglianoABViniciusLLahrMM 2007 Life history trade-offs explain the evolution of human pygmies. Proc Natl Acad Sci U S A 104 20216 20219
2. PerryGHDominyNJ 2009 Evolution of the human pygmy phenotype. Trends Ecol Evol 24 218 225
3. BatiniCLopesJBeharDMCalafellFJordeLB 2011 Insights into the demographic history of African Pygmies from complete mitochondrial genomes. Mol Biol Evol 28 1099 1110
4. Destro-BisolGCoiaVBoschiIVerginelliFCagliaA 2004 The analysis of variation of mtDNA hypervariable region 1 suggests that Eastern and Western Pygmies diverged before the Bantu expansion. Am Nat 163 212 226
5. PatinELavalGBarreiroLBSalasASeminoO 2009 Inferring the demographic history of African farmers and pygmy hunter-gatherers using a multilocus resequencing data set. PLoS Genet 5 e1000448 doi:10.1371/journal.pgen.1000448
6. Quintana-MurciLQuachHHarmantCLucaFMassonnetB 2008 Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter-gatherers and Bantu-speaking farmers. Proc Natl Acad Sci U S A 105 1596 1601
7. VerduPAusterlitzFEstoupAVitalisRGeorgesM 2009 Origins and genetic diversity of pygmy hunter-gatherers from Western Central Africa. Curr Biol 19 312 318
8. PhilipsonD 1975 The Chronology of the Iron Age in Bantu Africa. The Journal of African History 16 321 342
9. TishkoffSAReedFAFriedlaenderFREhretCRanciaroA 2009 The genetic structure and history of Africans and African Americans. Science 324 1035 1044
10. MerimeeTJZapfJHewlettBCavalli-SforzaLL 1987 Insulin-like growth factors in pygmies. The role of puberty in determining final stature. N Engl J Med 316 906 911
11. KangSJChiangCWPalmerCDTayoBOLettreG 2010 Genome-wide association of anthropometric traits in African - and African-derived populations. Hum Mol Genet 19 2725 2738
12. Lango AllenHEstradaKLettreGBerndtSIWeedonMN 2010 Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467 832 838
13. MerimeeTJRimoinDLCavalli-SforzaLL 1972 Metabolic studies in the African pygmy. J Clin Invest 51 395 401
14. RimoinDLMerimeeTJRabinowitzDCavalli-SforzaLLMcKusickVA 1969 Peripheral subresponsiveness to human growth hormone in the African pygmies. N Engl J Med 281 1383 1388
15. MerimeeTJZapfJFroeschER 1981 Dwarfism in the pygmy. An isolated deficiency of insulin-like growth factor I. N Engl J Med 305 965 968
16. JainSGoldeDWBaileyRGeffnerME 1998 Insulin-like growth factor-I resistance. Endocr Rev 19 625 646
17. HattoriYVeraJCRivasCIBerschNBaileyRC 1996 Decreased insulin-like growth factor I receptor expression and function in immortalized African Pygmy T cells. J Clin Endocrinol Metab 81 2257 2263
18. BaumannGShawMAMerimeeTJ 1989 Low levels of high-affinity growth hormone-binding protein in African pygmies. N Engl J Med 320 1705 1709
19. DavilaNSheaBTOmotoKMercadoMMisawaS 2002 Growth hormone binding protein, insulin-like growth factor-I and short stature in two pygmy populations from the Philippines. J Pediatr Endocrinol Metab 15 269 276
20. BozzolaMTravaglinoPMarzilianoNMeazzaCPaganiS 2009 The shortness of Pygmies is associated with severe under-expression of the growth hormone receptor. Mol Genet Metab 98 310 313
21. BeckerNSVerduPFromentALe BominSPagezyH 2011 Indirect evidence for the genetic determination of short stature in African Pygmies. Am J Phys Anthropol
22. WinklerCANelsonGWSmithMW 2010 Admixture mapping comes of age. Annu Rev Genomics Hum Genet 11 65 89
23. AkeyJM 2009 Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res 19 711 722
24. WeirBS 1996 Genetic data analysis II : methods for discrete population genetic data Sunderland, Mass. Sinauer Associates xii 445
25. ShriverMDKennedyGCParraEJLawsonHASonparV 2004 The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum Genomics 1 274 286
26. InnanHKimY 2008 Detecting local adaptation using the joint sampling of polymorphism data in the parental and derived populations. Genetics 179 1713 1720
27. LiJZAbsherDMTangHSouthwickAMCastoAM 2008 Worldwide human relationships inferred from genome-wide patterns of variation. Science 319 1100 1104
28. VoightBFKudaravalliSWenXPritchardJK 2006 A map of recent positive selection in the human genome. PLoS Biol 4 e72 doi:10.1371/journal.pbio.0040072
29. SabetiPCVarillyPFryBLohmuellerJHostetterE 2007 Genome-wide detection and characterization of positive selection in human populations. Nature 449 913 918
30. YoungLJWangZ 2004 The neurobiology of pair bonding. Nat Neurosci 7 1048 1054
31. KamathJYarbroughGGPrangeAJJrWinokurA 2009 The thyrotropin-releasing hormone (TRH)-immune system homeostatic hypothesis. Pharmacol Ther 121 20 28
32. DormitzerPREllisonPTBodeHH 1989 Anomalously low endemic goiter prevalence among Efe pygmies. Am J Phys Anthropol 78 527 531
33. Lopez HerraezDBauchetMTangKTheunertCPugachI 2009 Genetic variation and recent positive selection in worldwide human populations: evidence from nearly 1 million SNPs. PLoS ONE 4 e7888 doi:10.1371/journal.pone.0007888
34. EleftherohorinouHWrightVHoggartCHartikainenALJarvelinMR 2009 Pathway analysis of GWAS provides new insights into genetic susceptibility to 3 inflammatory diseases. PLoS ONE 4 e8068 doi:10.1371/journal.pone.0008068
35. PickrellJKCoopGNovembreJKudaravalliSLiJZ 2009 Signals of recent positive selection in a worldwide sample of human populations. Genome Res 19 826 837
36. La Batide-AlanoreATregouetDASassCSiestGVisvikisS 2003 Family study of the relationship between height and cardiovascular risk factors in the STANISLAS cohort. Int J Epidemiol 32 607 614
37. MooreLGShriverMBemisLHicklerBWilsonM 2004 Maternal adaptation to high-altitude pregnancy: an experiment of nature–a review. Placenta 25 Suppl A S60 71
38. MoorjaniPPattersonNHirschhornJNKeinanAHaoL 2011 The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet 7 e1001373 doi:10.1371/journal.pgen.1001373
39. PoolJENielsenR 2009 Inference of historical changes in migration rate from the lengths of migrant tracts. Genetics 181 711 719
40. PritchardJKDi RienzoA 2010 Adaptation - not by sweeps alone. Nat Rev Genet 11 665 667
41. KimuraRFujimotoATokunagaKOhashiJ 2007 A practical genome scan for population-specific strong selective sweeps that have reached fixation. PLoS ONE 2 e286 doi:10.1371/journal.pone.0000286
42. TangKThorntonKRStonekingM 2007 A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol 5 e171 doi:10.1371/journal.pbio.0050171
43. WilliamsonSHHubiszMJClarkAGPayseurBABustamanteCD 2007 Localizing recent adaptive evolution in the human genome. PLoS Genet 3 e90 doi:10.1371/journal.pgen.0030090
44. LanktreeMBGuoYMurtazaMGlessnerJTBaileySD 2011 Meta-analysis of Dense Genecentric Association Studies Reveals Common and Uncommon Variants Associated with Height. Am J Hum Genet 88 6 18
45. LudwigSEngelKHoffmeyerASithanandamGNeufeldB 1996 3pK, a novel mitogen-activated protein (MAP) kinase-activated protein kinase, is targeted by three MAP kinase pathways. Mol Cell Biol 16 6687 6697
46. AlexanderWSHiltonDJ 2004 The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu Rev Immunol 22 503 529
47. de SilvaMGElliottKDahlHHFitzpatrickEWilcoxS 2003 Disruption of a novel member of a sodium/hydrogen exchanger family and DOCK3 is associated with an attention deficit hyperactivity disorder-like phenotype. J Med Genet 40 733 740
48. KhorCCVannbergFOChapmanSJGuoHWongSH 2010 CISH and susceptibility to infectious diseases. N Engl J Med 362 2092 2101
49. YasukawaHSasakiAYoshimuraA 2000 Negative regulation of cytokine signaling pathways. Annu Rev Immunol 18 143 164
50. ChenYLinGHuoJSBarneyDWangZ 2009 Computational and functional analysis of growth hormone (GH)-regulated genes identifies the transcriptional repressor B-cell lymphoma 6 (Bc16) as a participant in GH-regulated transcription. Endocrinology 150 3645 3654
51. KiddJMCooperGMDonahueWFHaydenHSSampasN 2008 Mapping and sequencing of structural variation from eight human genomes. Nature 453 56 64
52. ChenJBillingsSENishimuneH 2011 Calcium channels link the muscle-derived synapse organizer laminin beta2 to Bassoon and CAST/Erc2 to organize presynaptic active zones. J Neurosci 31 512 525
53. JoronMFrezalLJonesRTChamberlainNLLeeSF 2011 Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477 203 206
54. RedelmanDWelniakLATaubDMurphyWJ 2008 Neuroendocrine hormones such as growth hormone and prolactin are integral members of the immunological cytokine network. Cell Immunol 252 111 121
55. BallardC 2006 Strange Alliance: Pygmies in the Colonial Imaginary. World Archaeology 38 133 151
56. LeonhardtA 2006 Baka and the Magic of the State: Between Autochthony and Citizenship. African Studies Review 49 69 94
57. PelicanM 2009 Complexities of Indigeneity and Autochthony: An African Example. American Ethnologist 36 52 65
58. MillerSADykesDDPoleskyHF 1988 A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16 1215
59. PurcellSNealeBTodd-BrownKThomasLFerreiraMA 2007 PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 559 575
60. Team RDC 2007 R: A language and environment for statistical computing Vienna R Foundation for Statistical Computing
61. PembertonTJWangCLiJZRosenbergNA 2010 Inference of unexpected genetic relatedness among individuals in HapMap Phase III. Am J Hum Genet 87 457 464
62. HuffCDHarpendingHCRogersAR 2010 Detecting positive selection from genome scans of linkage disequilibrium. BMC Genomics 11 8
63. ScheetPStephensM 2006 A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 78 629 644
64. McVeanGAMyersSRHuntSDeloukasPBentleyDR 2004 The fine-scale structure of recombination rate variation in the human genome. Science 304 581 584
65. 2005 A haplotype map of the human genome. Nature 437 1299 1320
66. SherrySTWardMHKholodovMBakerJPhanL 2001 dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29 308 311
67. FujitaPARheadBZweigASHinrichsASKarolchikD 2011 The UCSC Genome Browser database: update 2011. Nucleic Acids Res 39 D876 882
68. ChakrabortyRWeissKM 1988 Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc Natl Acad Sci U S A 85 9119 9123
69. SankararamanSSridharSKimmelGHalperinE 2008 Estimating local ancestry in admixed populations. Am J Hum Genet 82 290 303
70. PasaniucBSankararamanSKimmelGHalperinE 2009 Inference of locus-specific ancestry in closely related populations. Bioinformatics 25 i213 221
71. PriceALTandonAPattersonNBarnesKCRafaelsN 2009 Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet 5 e1000519 doi:10.1371/journal.pgen.1000519
72. BrycKAutonANelsonMROksenbergJRHauserSL 2010 Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci U S A 107 786 791
73. TangHSiegmundDOJohnsonNARomieuILondonSJ 2010 Joint testing of genotype and ancestry association in admixed families. Genet Epidemiol 34 783 791
74. BenjaminiYHochbergY 1995 Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 57 289 300
75. StoreyJDTibshiranaiR 2003 Statistical significance for genomewide studies. PNAS 100 9440 9445
76. KangHMSulJHServiceSKZaitlenNAKongSY 2010 Variance component model to account for sample structure in genome-wide association studies. Nat Genet 42 348 354
77. BoykoARQuignonPLiLSchoenebeckJJDegenhardtJD 2010 A simple genetic architecture underlies morphological variation in dogs. PLoS Biol 8 e1000451 doi:10.1371/journal.pbio.1000451
78. PriceALZaitlenNAReichDPattersonN 2010 New approaches to population stratification in genome-wide association studies. Nat Rev Genet 11 459 463
79. StephensMDonnellyP 2003 A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73 1162 1169
80. StephensMScheetP 2005 Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet 76 449 462
81. StephensMSmithNJDonnellyP 2001 A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68 978 989
82. BandeltHJForsterPRohlA 1999 Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16 37 48
83. LibradoPRozasJ 2009 DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25 1451 1452
Štítky
Genetika Reprodukční medicína
Článek A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target GenesČlánek Population Structure of Hispanics in the United States: The Multi-Ethnic Study of AtherosclerosisČlánek Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms inČlánek Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome AnalysisČlánek Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and MechanismČlánek Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 4
-
Všechny články tohoto čísla
- Runs of Homozygosity Implicate Autozygosity as a Schizophrenia Risk Factor
- Modifier Genes and the Plasticity of Genetic Networks in Mice
- The DSIF Subunits Spt4 and Spt5 Have Distinct Roles at Various Phases of Immunoglobulin Class Switch Recombination
- A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target Genes
- Population Structure of Hispanics in the United States: The Multi-Ethnic Study of Atherosclerosis
- Deep Sequencing of Plant and Animal DNA Contained within Traditional Chinese Medicines Reveals Legality Issues and Health Safety Concerns
- Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms in
- Insulin Signaling Mediates Sexual Attractiveness in
- Progressive Telomere Dysfunction Causes Cytokinesis Failure and Leads to the Accumulation of Polyploid Cells
- Long-Range Chromosome Organization in : A Site-Specific System Isolates the Ter Macrodomain
- Regulation of Budding Yeast Mating-Type Switching Donor Preference by the FHA Domain of Fkh1
- Polyglutamine Toxicity Is Controlled by Prion Composition and Gene Dosage in Yeast
- Patterns of Regulatory Variation in Diverse Human Populations
- Sequence-Specific Targeting of Dosage Compensation in Favors an Active Chromatin Context
- Whole-Exome Sequencing and Homozygosity Analysis Implicate Depolarization-Regulated Neuronal Genes in Autism
- Replication Fork Reversal after Replication–Transcription Collision
- Common Variants at 9p21 and 8q22 Are Associated with Increased Susceptibility to Optic Nerve Degeneration in Glaucoma
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Epigenome-Wide Scans Identify Differentially Methylated Regions for Age and Age-Related Phenotypes in a Healthy Ageing Population
- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Budding Yeast Dma Proteins Control Septin Dynamics and the Spindle Position Checkpoint by Promoting the Recruitment of the Elm1 Kinase to the Bud Neck
- , a Homolog of a Deaf-Blindness Gene, Regulates Circadian Output and Slowpoke Channels
- Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome Analysis
- Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and Mechanism
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
- SPE-44 Implements Sperm Cell Fate
- A Shared Role for RBF1 and dCAP-D3 in the Regulation of Transcription with Consequences for Innate Immunity
- A Companion Cell–Dominant and Developmentally Regulated H3K4 Demethylase Controls Flowering Time in via the Repression of Expression
- The HEN1 Ortholog, HENN-1, Methylates and Stabilizes Select Subclasses of Germline Small RNAs
- Improved Statistics for Genome-Wide Interaction Analysis
- The Probability of a Gene Tree Topology within a Phylogenetic Network with Applications to Hybridization Detection
- Context-Dependent Dual Role of SKI8 Homologs in mRNA Synthesis and Turnover
- Mu Insertions Are Repaired by the Double-Strand Break Repair Pathway of
- Competition between Replicative and Translesion Polymerases during Homologous Recombination Repair in Drosophila
- An Unbiased Assessment of the Role of Imprinted Genes in an Intergenerational Model of Developmental Programming
- Type 2 Diabetes Risk Alleles Demonstrate Extreme Directional Differentiation among Human Populations, Compared to Other Diseases
- Mutations in and Cause “Splashed White” and Other White Spotting Phenotypes in Horses
- Fine-Scale Mapping of Natural Variation in Fly Fecundity Identifies Neuronal Domain of Expression and Function of an Aquaporin
- Dynamics of Brassinosteroid Response Modulated by Negative Regulator LIC in Rice
- Genetic Inhibition of Solute-Linked Carrier 39 Family Transporter 1 Ameliorates Aβ Pathology in a Model of Alzheimer's Disease
- The Functions of Mediator in Support a Role in Shaping Species-Specific Gene Expression
- Patterns of Ancestry, Signatures of Natural Selection, and Genetic Association with Stature in Western African Pygmies
- Dissection of Pol II Trigger Loop Function and Pol II Activity–Dependent Control of Start Site Selection
- PIWI Associated siRNAs and piRNAs Specifically Require the HEN1 Ortholog
- Genome-Wide Patterns of Gene Expression in Nature
- Hypoxia Disruption of Vertebrate CNS Pathfinding through EphrinB2 Is Rescued by Magnesium
- A New Role for Translation Initiation Factor 2 in Maintaining Genome Integrity
- Sex Reversal in C57BL/6J XY Mice Caused by Increased Expression of Ovarian Genes and Insufficient Activation of the Testis Determining Pathway
- The Rac GTP Exchange Factor TIAM-1 Acts with CDC-42 and the Guidance Receptor UNC-40/DCC in Neuronal Protrusion and Axon Guidance
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



