-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaGenetics of Ribosomal Proteins: “Curiouser and Curiouser”
article has not abstract
Published in the journal: . PLoS Genet 9(1): e32767. doi:10.1371/journal.pgen.1003300
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1003300Summary
article has not abstract
The Mystery of Minutes
Lewis Carroll wrote about Alice, but he might just as well have been referring to Calvin Bridges. As a student in T.H. Morgan's lab, Bridges described some of the earliest Drosophila mutations, including so-called Minutes, in which heterozygotes exhibited small body size and developmental abnormalities in tissues undergoing rapid cell division, and homozygotes were lethal [1]. At the time, it was curious how dozens of different loci could yield the same phenotype, and even curiouser how flies multiply heterozygous at different Minute loci were no more severely affected than a single Minute mutant. This mystery—how dozens of genes could encode similar but separate proliferative functions in all cells—was solved more than 50 years later with the realization that mutations of ribosomal protein genes occur in almost all Minute loci [2].
In this issue of PLOS Genetics, Watkins-Chow et al. [1] add to a more recent curiosity: even though ribosomes (and ribosomal protein [RP] genes) have remained nearly identical across more than a billion years of evolution, mutations of RP genes in mice and in humans give rise to a surprising diversity of phenotypes. This work adds a new piece to a very old puzzle, and suggests the possibility that RP genes do more than just contribute to ribosomes.
RP Mutations in Mammals: More than Minutes
To date, 11 different RP mutant mice have now been reported. These mice carry deletions, missense, or splicing mutations that have arisen spontaneously, from N-Ethyl-N-Nitrosurea (ENU) mutagenesis screens, or from targeted gene deletion [1]–[11] (Table 1). Overall, RP mutant mice exhibit an unexpected array and diversity of phenotypes. For example, the spontaneous mutant Belly spot and tail is caused by a splicing abnormality in Rpl24 [6]; heterozygous mutants (Rpl24Bst/+) are small with white hind feet, a midline belly spot (Bst), abnormal retinal development, and skeletal abnormalities that include a curly tail [4]. Heterozygosity for a targeted mutation of Rps6 causes embryonic lethality [12], while heterozygosity for targeted mutations of Rpl22 or of Rpl29 have no effect. (Homozygosity for targeted mutations of Rpl22 and of Rpl29 causes a T-cell–specific developmental defect [6] and generalized reduced growth [8], respectively.) Mice heterozygous for mutations in Rps19, Rps20, or Rpl27a exhibit epidermal hyperpigmentation, anemia, and reduced body size while the Rpl27a heterozygotes also exhibit cerebellar ataxia. Rps19-null mice are embryonic lethal prior to implantation [10]. CD74-Nid67+/− mice with a heterozygous deletion of eight genes including Rps14 developed macrocytic anemia and other hematopoietic defects [11]. Finally, heterozygous Rpl38 mutants had abnormalities of skeletal patterning [9].
Tab. 1. Mouse Models of Ribosomal Proteins. 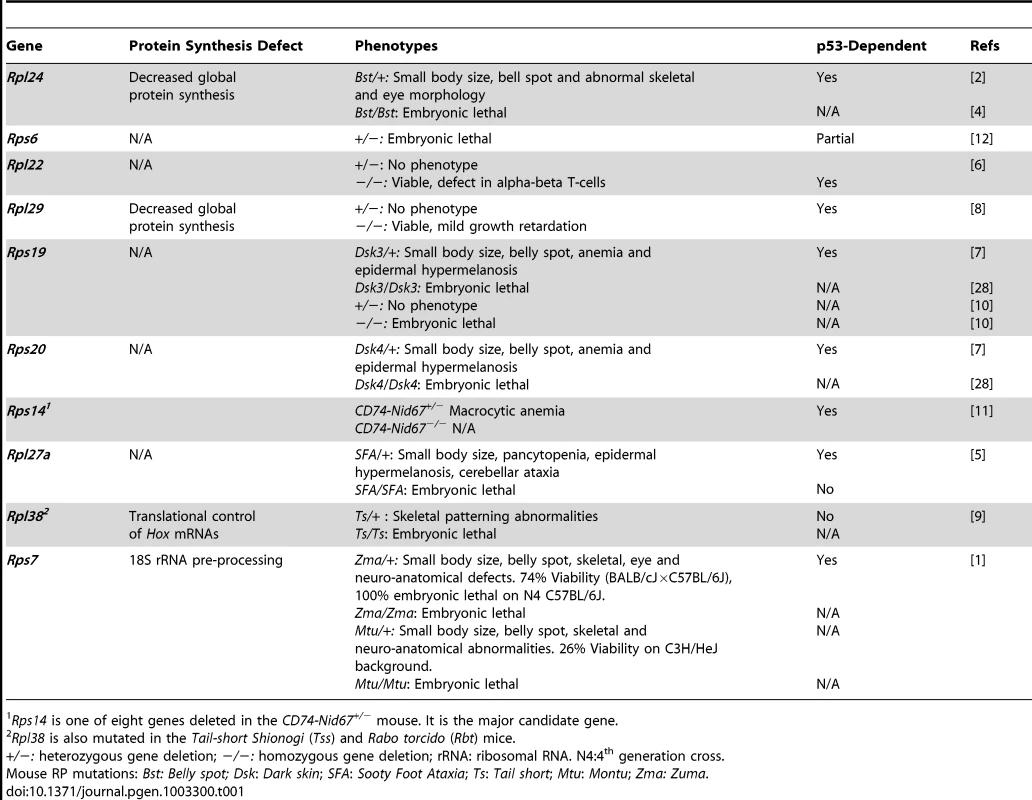
Rps14 is one of eight genes deleted in the CD74-Nid67+/− mouse. It is the major candidate gene. Watkins-Chow et al. [1] present two different missense alleles of Rps7, Montu (Mtu) and Zuma (Zma), that were generated from an ENU mutagenesis screen. Most Rps7Mtu/+ mice die in utero (74% on a C3H/HeJ background), but the survivors show pleiotropic phenotypes including reduced body size, abnormal skeletal morphology, and mid-ventral white spotting. This phenotype cluster is reproduced in Rps7Zma/+ mice and is similar to that in Rpl24 mutant mice [1]. These observations support the existence of distinctive spatial and temporal characteristics for RPs [13]: Rps7, Rps19, Rps20, and Rpl24 may be necessary for melanocyte development [1], [2], [4], [7], while Rps6, Rps19, Rps20, and Rpl27a are important for keratinocytes [3], [7]. Additionally, Rps7, Rpl24, and Rpl38 are crucial for skeletal and retinal development [1], [2], [9] and Rps14, Rps19, Rps20, and Rpl27a are necessary for hematopoeisis [3], [7], [11]. RP genotype–phenotype correlations are also found in humans where mutations cause diseases with similarly complex clinical manifestations such as Diamond Blackfan anemia (DBA). DBA is characterized by diverse abnormalities including anemia, congenital craniofacial malformations, and defects in kidney development [14].
The Role of p53 in RP Haploinsufficiency Phenotypes
While disturbances in ribosome biogenesis have been linked to many human diseases [15], there is also increasing evidence that RP mutations may be associated with cancer susceptibility [15]–[17]. In keeping with these observations, a sensitive connection between RPs and the tumor suppressor p53 has been identified (reviewed in [18]). p53 is a major cellular stress sensor that is best known for its ability to induce apoptosis, cell cycle arrest, and senescence in response to a variety of insults including DNA damage, oncogene activation, hypoxia, and more recently, ribosomal stress [19]. Thus, the p53 pathway provides a surveillance mechanism for the preservation of genomic and ribosomal integrity. Perturbation of ribosomal integrity induces the release of several RPs from the nucleolus that interact with and suppress the activity of the main negative regulator of p53, Mdm2, leading to the stabilization and activation of p53 [20]–[25]. This is observed in a number of RP mutant mice that exhibit cell cycle arrest and apoptosis in the affected cell types and in which the resulting pathologies are ameliorated or suppressed by p53 deletion (Table 1). In Rps19Dsk3/+, Rps20Dsk4/+, Rpl24Bst/+, Rpl27aSfa/+, and Rps7Zma/+ mice, the removal of one p53 copy is sufficient to alleviate all phenotypic abnormalities [1]–[3], [7]. In the case of Rps6+/− mice, fetal mortality is delayed by only a couple of days in the absence of p53 [12]. Amazingly, the embryonic lethality of Rps7Zma/+ mice was completely suppressed by the loss of one p53 allele, and Rps7Zma/+:p53+/− mice are for the most part identical to their wild-type littermates [1]. Taken together, these experiments establish p53 as a true sensor of nucleolar stress and highlight the extraribosomal activity of RPs as modulators of the Mdm2–p53 pathway.
RP Functions in Mutant Mice
Mutant RP phenotypes in mice appear to be the result of three distinct mechanisms: 1) global suppression of protein synthesis; 2) specific suppression of protein synthesis; and 3) extra-ribosomal functions. Diminished global protein synthesis was identified in Rpl24Bst/+ (∼30% reduced) and Rpl29−/− mouse neural tube and somites (∼45% reduced), although no concurrent phenotype was described [9]. In Rps7Mtu/+ mice, a novel defect in 18S rRNA preprocessing was identified in brain and liver, without a reduction in protein synthesis. We speculate that an accompanying reduction in protein synthesis could explain the homozygous embryonic lethality in these mice. On the other hand, the abnormal homeotic transformations observed in Rpl38Ts/+ mice were attributed to the unique role of Rpl38 in translation of specific Hox mRNAs rather than to a global effect [9]. RPL38 appeared to facilitate 80S complex formation during the earliest steps of translation initiation on selective Hox mRNAs. In addition to these global and gene-specific translational defects, extra-ribosomal functions of numerous RPs have been described in prokaryotes and lower eukaryotes such as yeast and flies, and in cultured human cells [13], [26], [27]. This expanding list of functions includes cellular apoptosis, transcription and mRNA processing, DNA repair, development, and tumorigenesis. To date, the ability of RPs to activate p53 is the only described extra-ribosomal function in mice. In Rps7Mtu/+ mice, impaired rRNA preprocessing and p53 activation occur simultaneously, illustrating the complex roles of RPs in mammalian tissues. With this in mind, mouse RP phenotypes (heterozygous and homozygous) need to be analyzed on a p53-null as well as on a p53 wild-type genetic background. Since so little is known about how changes in ribosomal protein levels may impact cellular function in vivo, a full repertoire of mouse RP models and improved characterization are sorely needed.
Due to the essential nature of RPs, it is apparent that even small perturbations to their functions can result in an increasing array of diseases, and it is clear now more than ever that the mystery of the underlying mechanistic basis for RP mutant phenotypes needs to be solved before state-of-the art translational approaches can bring effective treatments. Moreover, additional mouse models for RP disorders will provide an important preclinical resource for developing new treatments of ribosomopathies.
Zdroje
1. Watkins-ChowDE, CookeJ, PidsleyR, EdwardsA, SlotkinR, et al. (2013) Mutation of the Diamond-Blackfan anemia gene Rps7 in mouse results in morphological and neuroanatomical phenotypes. PLoS Genet 9: e1003094 doi:10.1371/journal.pgen.1003094.
2. BarkicM, CrnomarkovicS, GrabusicK, BogeticI, PanicL, et al. (2009) The p53 tumor suppressor causes congenital malformations in Rpl24-deficient mice and promotes their survival. Mol Cell Biol 29 : 2489–2504.
3. TerzianT, DumbleM, ArbabF, ThallerC, DonehowerLA, et al. (2011) Rpl27a mutation in the sooty foot ataxia mouse phenocopies high p53 mouse models. J Pathol 224 : 540–552.
4. OliverER, SaundersTL, TarleSA, GlaserT (2004) Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development 131 : 3907–3920.
5. VolarevicS, StewartMJ, LedermannB, ZilbermanF, TerraccianoL, et al. (2000) Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science 288 : 2045–2047.
6. AndersonSJ, LauritsenJP, HartmanMG, FousheeAM, LefebvreJM, et al. (2007) Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity 26 : 759–772.
7. McGowanKA, LiJZ, ParkCY, BeaudryV, TaborHK, et al. (2008) Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet 40 : 963–970.
8. Kirn-SafranCB, OristianDS, FochtRJ, ParkerSG, VivianJL, et al. (2007) Global growth deficiencies in mice lacking the ribosomal protein HIP/RPL29. Dev Dyn 236 : 447–460.
9. KondrashovN, PusicA, StumpfCR, ShimizuK, HsiehAC, et al. (2011) Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145 : 383–397.
10. MatssonH, DaveyEJ, DraptchinskaiaN, HamaguchiI, OokaA, et al. (2004) Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Mol Cell Biol 24 : 4032–4037.
11. BarlowJL, DrynanLF, HewettDR, HolmesLR, Lorenzo-AbaldeS, et al. (2010) A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q - syndrome. Nat Med 16 : 59–66.
12. PanicL, TamarutS, Sticker-JantscheffM, BarkicM, SolterD, et al. (2006) Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol Cell Biol 26 : 8880–8891.
13. WarnerJR, McIntoshKB (2009) How common are extraribosomal functions of ribosomal proteins? Mol Cell 34 : 3–11.
14. BoriaI, GarelliE, GazdaHT, AspesiA, QuarelloP, et al. The ribosomal basis of Diamond-Blackfan Anemia: mutation and database update. Hum Mutat 31 : 1269–1279.
15. NarlaA, EbertBL Ribosomopathies: human disorders of ribosome dysfunction. Blood 115 : 3196–3205.
16. MontanaroL, TrereD, DerenziniM (2008) Nucleolus, ribosomes, and cancer. Am J Pathol 173 : 301–310.
17. RaoS, LeeSY, GutierrezA, PerrigoueJ, ThapaRJ, et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood 120 : 3764–3773.
18. ChakrabortyA, UechiT, KenmochiN Guarding the ‘translation apparatus’: defective ribosome biogenesis and the p53 signaling pathway. Wiley Interdiscip Rev RNA 2 : 507–522.
19. KruseJP, GuW (2009) Modes of p53 regulation. Cell 137 : 609–622.
20. DaiMS, ZengSX, JinY, SunXX, DavidL, et al. (2004) Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol 24 : 7654–7668.
21. DaiMS, LuH (2004) Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem 279 : 44475–44482.
22. LohrumMA, LudwigRL, KubbutatMH, HanlonM, VousdenKH (2003) Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3 : 577–587.
23. ZhangY, WolfGW, BhatK, JinA, AllioT, et al. (2003) Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 23 : 8902–8912.
24. ChenD, ZhangZ, LiM, WangW, LiY, et al. (2007) Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 26 : 5029–5037.
25. ZhuY, PoyurovskyMV, LiY, BidermanL, StahlJ, et al. (2009) Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell 35 : 316–326.
26. WoolIG (1996) Extraribosomal functions of ribosomal proteins. Trends Biochem Sci 21 : 164–165.
27. LindstromMS (2009) Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem Biophys Res Commun 379 : 167–170.
28. FitchKR, McGowanKA, van RaamsdonkCD, FuchsH, LeeD, et al. (2003) Genetics of dark skin in mice. Genes Dev 17 : 214–228.
Štítky
Genetika Reprodukční medicína
Článek Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across PathogensČlánek TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association StudiesČlánek Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization inČlánek Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA ExpressionČlánek The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of GenesČlánek The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 1
-
Všechny články tohoto čísla
- A Model of High Sugar Diet-Induced Cardiomyopathy
- Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across Pathogens
- Emerging Function of Fat Mass and Obesity-Associated Protein (Fto)
- Positional Cloning Reveals Strain-Dependent Expression of to Alter Susceptibility to Bleomycin-Induced Pulmonary Fibrosis in Mice
- Genetics of Ribosomal Proteins: “Curiouser and Curiouser”
- Transposable Elements Re-Wire and Fine-Tune the Transcriptome
- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- MAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
- Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies
- A Systematic Mapping Approach of 16q12.2/ and BMI in More Than 20,000 African Americans Narrows in on the Underlying Functional Variation: Results from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
- Histone H3K56 Acetylation, Rad52, and Non-DNA Repair Factors Control Double-Strand Break Repair Choice with the Sister Chromatid
- Genome-Wide Association Study Identifies a Novel Susceptibility Locus at 12q23.1 for Lung Squamous Cell Carcinoma in Han Chinese
- Genetic Disruption of the Copulatory Plug in Mice Leads to Severely Reduced Fertility
- The [] Prion Exists as a Dynamic Cloud of Variants
- Adult Onset Global Loss of the Gene Alters Body Composition and Metabolism in the Mouse
- Fis Protein Insulates the Gene from Uncontrolled Transcription
- The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse
- Genome-Wide Haplotype Analysis of Expression Quantitative Trait Loci in Monocytes
- TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association Studies
- Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
- The Ecm11-Gmc2 Complex Promotes Synaptonemal Complex Formation through Assembly of Transverse Filaments in Budding Yeast
- MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
- Comparative Genomic Analysis of the Endosymbionts of Herbivorous Insects Reveals Eco-Environmental Adaptations: Biotechnology Applications
- Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry
- Pharmacodynamics, Population Dynamics, and the Evolution of Persistence in
- A Hybrid Likelihood Model for Sequence-Based Disease Association Studies
- Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity
- Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis
- Transcriptional Dynamics Elicited by a Short Pulse of Notch Activation Involves Feed-Forward Regulation by Genes
- Dynamic Large-Scale Chromosomal Rearrangements Fuel Rapid Adaptation in Yeast Populations
- Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability
- Gene Copy-Number Polymorphism Caused by Retrotransposition in Humans
- An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in
- Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization in
- Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination
- Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes and
- Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene
- A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
- Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators
- Genome-Wide Analysis Reveals Selection for Important Traits in Domestic Horse Breeds
- Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
- Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
- HIF- and Non-HIF-Regulated Hypoxic Responses Require the Estrogen-Related Receptor in
- Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages
- The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Loci Associated with -Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers
- Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol
- Centromere-Like Regions in the Budding Yeast Genome
- Sequencing of Loci from the Elephant Shark Reveals a Family of Genes in Vertebrate Genomes, Forged by Ancient Duplications and Divergences
- Mendelian and Non-Mendelian Regulation of Gene Expression in Maize
- Mutational Spectrum Drives the Rise of Mutator Bacteria
- Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression
- The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of Genes
- Sex-Specific Signaling in the Blood–Brain Barrier Is Required for Male Courtship in
- A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity
- Is Required for Leptin-Mediated Depolarization of POMC Neurons in the Hypothalamic Arcuate Nucleus in Mice
- Unlocking the Bottleneck in Forward Genetics Using Whole-Genome Sequencing and Identity by Descent to Isolate Causative Mutations
- The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
- MTERF3 Regulates Mitochondrial Ribosome Biogenesis in Invertebrates and Mammals
- Downregulation and Altered Splicing by in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
- NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses
- Retroactive Maintains Cuticle Integrity by Promoting the Trafficking of Knickkopf into the Procuticle of
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network
- Genetic and Functional Modularity of Activities in the Specification of Limb-Innervating Motor Neurons
- A Population Genetic Model for the Maintenance of R2 Retrotransposons in rRNA Gene Loci
- A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in
- A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1)
- Mutation of the Diamond-Blackfan Anemia Gene in Mouse Results in Morphological and Neuroanatomical Phenotypes
- Life, the Universe, and Everything: An Interview with David Haussler
- Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome Oxidase Blockade and Limits Mitochondrial ROS Overproduction
- An Evolutionarily Conserved Synthetic Lethal Interaction Network Identifies FEN1 as a Broad-Spectrum Target for Anticancer Therapeutic Development
- The Flowering Repressor Underlies a Novel QTL Interacting with the Genetic Background
- Telomerase Is Required for Zebrafish Lifespan
- and Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
- WNK1/HSN2 Mutation in Human Peripheral Neuropathy Deregulates Expression and Posterior Lateral Line Development in Zebrafish ()
- Synergistic Interaction of Rnf8 and p53 in the Protection against Genomic Instability and Tumorigenesis
- Dot1-Dependent Histone H3K79 Methylation Promotes Activation of the Mek1 Meiotic Checkpoint Effector Kinase by Regulating the Hop1 Adaptor
- A Heterogeneous Mixture of F-Series Prostaglandins Promotes Sperm Guidance in the Reproductive Tract
- Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
- Directed Evolution of a Model Primordial Enzyme Provides Insights into the Development of the Genetic Code
- Genome-Wide Screens for Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



