-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Reklamaand Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
Termination of the stem cells in the floral meristem (also known as floral determinacy) is critical for the reproductive success of plants, and the molecular activities regulating floral determinacy are precisely orchestrated during the course of floral development. In Arabidopsis thaliana, regulators of floral determinacy include several transcription factor genes, such as APETALA2 (AP2), AGAMOUS (AG), SUPERMAN (SUP), and CRABSCLAW (CRC), as well as a microRNA (miRNA), miR172, which targets AP2. How the transcription factor and miRNA genes are coordinately regulated to achieve floral determinacy is unknown. A mutation in POWERDRESS (PWR), a previously uncharacterized gene encoding a SANT-domain-containing protein, was isolated in this study as an enhancer of the weakly indeterminate ag-10 allele. PWR was found to promote the transcription of CRC, MIR172a, b, and c and/or enhance Pol II occupancy at their promoters, without affecting MIR172d or e. A mutation in mature miR172d was additionally found to enhance the determinacy defects of ag-10 in an AP2-dependent manner, providing direct evidence that miR172d is functional in repressing AP2 and thereby contributes to floral determinacy. Thus, while PWR promotes floral determinacy by enhancing the expression of three of the five MIR172 members as well as CRC, MIR172d, whose expression is PWR-independent, also functions in floral stem cell termination. Taken together, these findings demonstrate how transcriptional diversification and functional redundancy of a miRNA family along with PWR-mediated co-regulation of miRNA and transcription factor genes contribute to the robustness of the floral determinacy network.
Published in the journal: . PLoS Genet 9(1): e32767. doi:10.1371/journal.pgen.1003218
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003218Summary
Termination of the stem cells in the floral meristem (also known as floral determinacy) is critical for the reproductive success of plants, and the molecular activities regulating floral determinacy are precisely orchestrated during the course of floral development. In Arabidopsis thaliana, regulators of floral determinacy include several transcription factor genes, such as APETALA2 (AP2), AGAMOUS (AG), SUPERMAN (SUP), and CRABSCLAW (CRC), as well as a microRNA (miRNA), miR172, which targets AP2. How the transcription factor and miRNA genes are coordinately regulated to achieve floral determinacy is unknown. A mutation in POWERDRESS (PWR), a previously uncharacterized gene encoding a SANT-domain-containing protein, was isolated in this study as an enhancer of the weakly indeterminate ag-10 allele. PWR was found to promote the transcription of CRC, MIR172a, b, and c and/or enhance Pol II occupancy at their promoters, without affecting MIR172d or e. A mutation in mature miR172d was additionally found to enhance the determinacy defects of ag-10 in an AP2-dependent manner, providing direct evidence that miR172d is functional in repressing AP2 and thereby contributes to floral determinacy. Thus, while PWR promotes floral determinacy by enhancing the expression of three of the five MIR172 members as well as CRC, MIR172d, whose expression is PWR-independent, also functions in floral stem cell termination. Taken together, these findings demonstrate how transcriptional diversification and functional redundancy of a miRNA family along with PWR-mediated co-regulation of miRNA and transcription factor genes contribute to the robustness of the floral determinacy network.
Introduction
Flowers are a key adaptation of the angiosperm lineage that helps ensure reproductive success. At a particular point in floral development, stem cells in the floral meristem are terminated, thereby preventing the continual growth that typifies the shoot and root apical meristems of plants. In Arabidopsis thaliana, the floral homeotic gene AGAMOUS (AG) is required for both reproductive organ specification and floral determinacy. AG's role in floral determinacy involves the negative regulation of the stem-cell-promoting gene WUSCHEL (WUS) in the floral meristem, via direct and indirect mechanisms [1]–[3].
Other known regulators of floral determinacy represent a wide range of functions and include signaling proteins, transcription factors, and microRNAs. Of particular importance for the present study are the YABBY transcription factor CRABSCLAW (CRC) and the floral homeotic transcription factor APETALA2 (AP2). CRC's dual functions in determinacy and carpel development are notably similar to those of AG, and the subtly indeterminate crc-1 mutant background has helped uncover the involvement of a number of other genes in floral determinacy [4]–[6]. AP2 encodes an AP2-domain-containing transcription factor and is regulated by miR172. A role for AP2 in maintaining stem cell fate was first proposed in the shoot apical meristem [7]. A similar role in the floral meristem was revealed when miR172-resistant AP2 was expressed and found to confer indeterminate floral organ production [8]. This finding also suggested an indirect role for miR172, which limits the accumulation of AP2 protein by translational inhibition [9], in negatively regulating WUS to confer floral determinacy.
miRNAs, regulatory molecules 20–24 nucleotide in length, play critical roles in various aspects of plant development through sequence-specific regulation of their targets [10]–[12]. In plants, genes encoding miRNAs (MIR genes) typically reside between protein-coding genes and are transcribed by RNA polymerase II (Pol II) [13], [14]. Deeply conserved plant MIR genes typically belong to families with multiple members [15], [16]. Evidence from several MIR gene families in plants indicates that their evolution has features in common with the evolution of protein-coding gene families. For example, analysis of the MIR156, MIR159, and MIR166 families revealed differences in the spatial and temporal expression of genes within these families, which suggests that expression diversification occurred after gene duplication [17]. What is less clear, however, is how diversification in the expression of individual family members specifically contributes to the developmental processes regulated by the mature miRNA species produced by a given family.
miR172 regulates AP2 transcription factor genes involved in the distinct processes of flowering time and floral development [18], but the individual contributions of the five MIR172 genes to these processes are unknown. Three unique mature miR172 sequences are produced from these five loci (one from MIR172a and b, one from MIR172c and d, and one from MIR172e), and publicly available datasets indicate that the three mature miR172 species differentially accumulate in inflorescences, rosette leaves, seedlings, and siliques [19]. Furthermore, specific regulation of MIR172c and MIR172e in the outermost floral whorl by a transcriptional corepressor indicates that the MIR172 family members are differentially regulated in the flower [9], [20], [21].
The complex interplay of both MIR172 and transcription factor genes in the termination of the floral stem cells makes this developmental paradigm well suited for the investigation of how individual MIR family members contribute to the functions broadly assigned to the mature miRNA species. In the present study, the POWERDRESS (PWR) gene was found to promote floral determinacy through CRC and miR172. PWR promoted the transcription and/or enhanced Pol II occupancy at the promoters of MIR172a, b, and c but did not affect MIR172d or e. Coincidentally, a mutation in MIR172d was also found to enhance the determinacy defects of a weak ag allele. Taken together, these findings indicate that differential regulation of MIR172 genes with overlapping functions enhances the robustness of the genetic network underlying floral stem cell termination.
Results
PWR is required for the proper termination of floral stem cells
To identify genes that regulate the termination of the stem cells in the floral meristem, an ethyl methanesulfonate screen was performed in the ag-10 background, as previously reported [3], [22]. In contrast to the null ag-1 mutant, which produces sterile flowers that indeterminately produce sepals and petals [23], the weak ag-10 mutant has normal floral organ specification and slightly bulged gynoecia that very rarely, if at all, contain additional floral organs inside (Figure 1C, 1H; Table 1) [22]. Typical genetic enhancers isolated from the ag-10 screen included those with consistently bulged and shortened siliques. In one of these mutants, elongated gynophores (i.e., the structure supporting the gynoecium) were observed along with the presence of ectopic floral organs in all of the dissected gynoecia, indicating a consistent enhancement of the floral determinacy defect of ag-10 (Figure 1D, 1H; Table 1). The mutant also showed a small but statistically significant increase in floral organ number in the inner two whorls (Table 1). Map-based cloning revealed a G-to-A mutation that introduced a premature stop codon at the 372nd residue of a previously uncharacterized gene, At3g52250 (Figure 1A). The gene model for At3g52250 predicts a 1,656 amino acid protein with two SANT/Myb domains and putative DNA-binding and transcription factor activity (http://arabidopsis.org). SANT domains have high structural similarity to Myb DNA-binding domains but have been characterized as histone-binding domains important for chromatin remodeling activity in both plants and animals [24], [25]. The gene was subsequently named POWERDRESS (PWR) (based on the single mutant phenotype, described below), and the ag-10 enhancer mutation was designated pwr-1. The floral determinacy defects of ag-10 pwr-1 were rescued by transformation with an 8.1-kb genomic PWR fragment. As with ag-10 plants, the flowers from five independent pPWR:PWR-GFP lines in the ag-10 pwr-1 background rarely produced gynoecia containing ectopic floral organs, and elongated gynophores were not observed (Figure 1J).
Fig. 1. Gene diagram of PWR and phenotypes of ag-10, ag-10 pwr-1, ag-10Col, and ag-10Col pwr-2. 
(A) Schematic diagram indicating the position of the G-to-A mutation in pwr-1 that produces a premature stop codon and the approximate location of the T-DNA insertion in pwr-2. The regions encoding the putative SANT domains are also indicated. (B) Wild-type Ler flower. (C) ag-10 flower with slightly bulged carpels. (D) ag-10 pwr-1 flower with narrow, slightly folded petals and a shortened and bulged gynoecium. (E) Wild-type Col flower. (F) ag-10Col flower. Col and ag-10Col flowers are indistinguishable. (G) ag-10Col pwr-2 flower with slightly aberrant carpels. (H) From left to right, siliques of Ler, ag-10, and ag-10 pwr-1. ag-10 pwr-1 has visible gynophores bearing shortened and bulged gynoecia containing internal flowers. (I) From left to right, siliques of Col, ag-10Col, and ag-10Col pwr-2. Although most ag-10Col pwr-2 siliques have very subtle carpel defects, some are visibly bulged and contain additional floral organs inside. (J) Rescue analysis for ag-10 pwr-1. From left to right, ag-10, ag-10 pwr-1, and pPWR:PWR-GFP in the ag-10 pwr-1 background. Scale bars = 500 bp in (A), 5 mm in (H) to (J), and 1 mm in all other panels. Tab. 1. Floral organ counting and analysis of bulged carpel phenotypes. 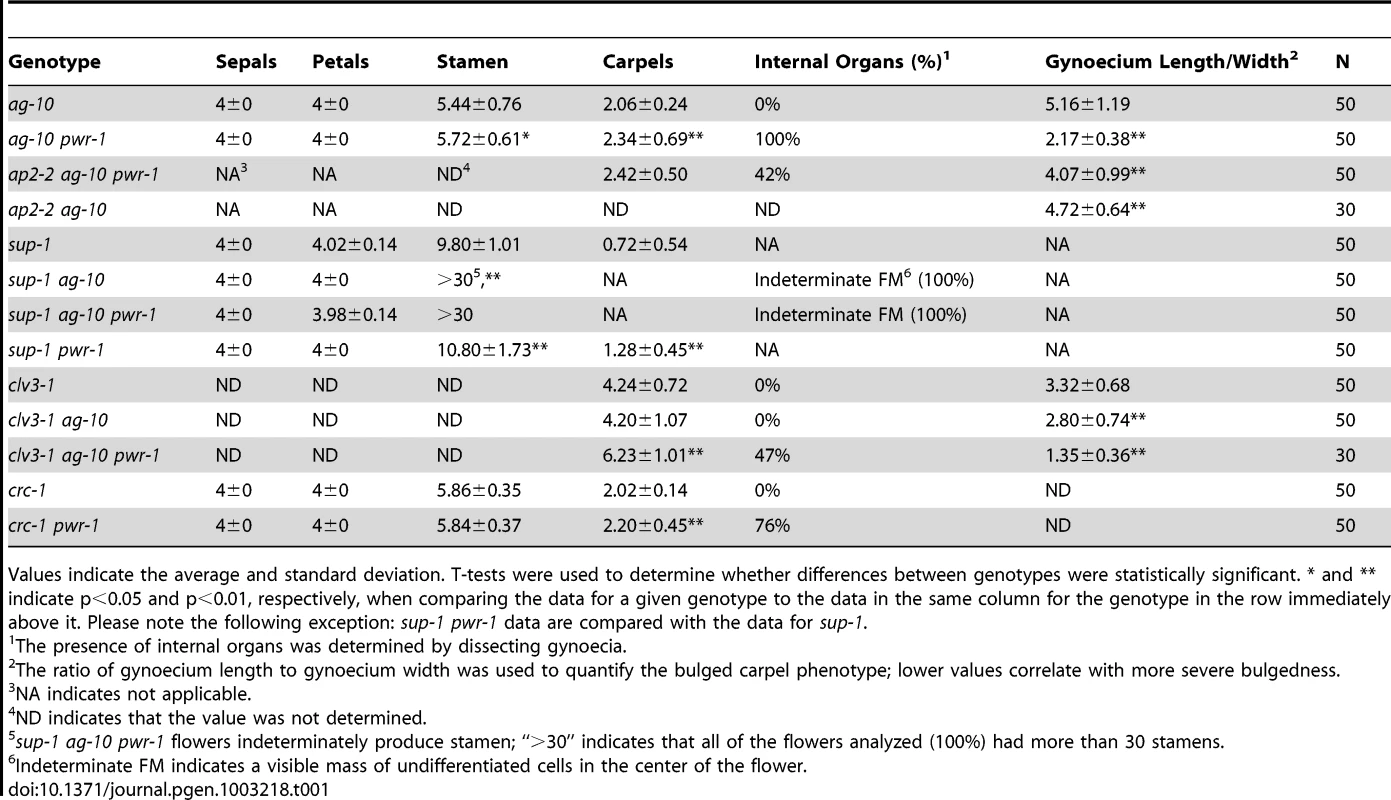
Values indicate the average and standard deviation. T-tests were used to determine whether differences between genotypes were statistically significant. * and ** indicate p<0.05 and p<0.01, respectively, when comparing the data for a given genotype to the data in the same column for the genotype in the row immediately above it. Please note the following exception: sup-1 pwr-1 data are compared with the data for sup-1. In addition to the rescue analysis, a T-DNA insertion line in the Columbia (Col) background, SALK_071811C (hereafter referred to as pwr-2), was obtained from the Arabidopsis Biological Resource Center [26]. In this mutant, the T-DNA insertion is near the 3′ end of the second exon of PWR (Figure 1A). To determine whether pwr-2 also affects floral determinacy, pwr-2 was crossed to ag-10Col, a line in which the ag-10 mutation was backcrossed into Col six times. Unlike ag-10 in the Landsberg erecta (Ler) background, ag-10Col siliques are nearly indistinguishable from wild-type siliques, indicating the presence of a genetic suppressor of determinacy defects in Col [3]. ag-10Col pwr-2 double-mutant flowers were found to exhibit weak determinacy defects, namely, the infrequent production of visibly bulged gynoecia containing additional floral organs inside (Figure 1I). These findings and the ag-10 pwr-1 double mutant phenotype indicate that PWR contributes to floral stem cell termination.
PWR impacts other developmental processes
Both ag-10 pwr-1 and ag-10Col pwr-2 mutants exhibited pleiotropic defects in addition to those in floral determinacy, including reduced plant height, early flowering, and aberrant petal shape. Although the defects were generally more pronounced in the Ler ecotype (e.g., ag-10 pwr-1 petals [Figure 1D] versus ag-10Col pwr-2 petals [Figure 1G]), pleiotropic phenotypes were observed in both the pwr-1 and pwr-2 single mutants. Defects in floral determinacy were not observed in the single mutants, but carpel development was visibly affected in both pwr alleles. pwr-1 and pwr-2 siliques were slightly flattened with the carpels bulged at the tip surrounding the stigmatic tissue (Figure 2A, 2B; the name POWERDRESS refers to the resemblance of the bulged carpel tips of the single mutants to the excessively padded shoulders of suit jackets from the 1980s).
Fig. 2. Phenotypes of pwr-1 and pwr-2 and transcript levels of genes involved in floral development. 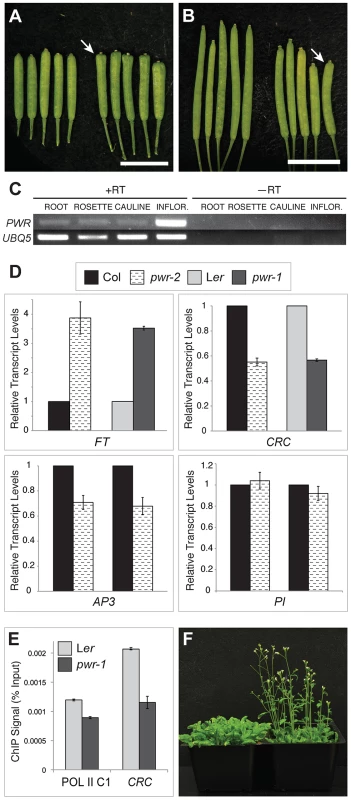
(A) Ler (left) and pwr-1 (right) siliques. (B) Col (left) and pwr-2 (right) siliques. In (A) and (B), arrows indicate the bulged carpel tips of pwr-1 and pwr-2 siliques, and scale bars = 5 mm. (C) RT-PCR for PWR using cDNA obtained from Ler roots, rosette leaves, cauline leaves, and inflorescences. UBIQUITIN 5 was used as the control. The highest abundance of the PWR transcript was observed in inflorescences. (D) Real-time RT-PCR analysis of the transcript levels of AP3, PI, CRC, and FT. For AP3 and PI, two biological replicates are shown for pwr-2 compared to Col. For CRC and FT, data is shown for both pwr alleles. For all genes tested, transcript levels were normalized to UBIQUITIN 5 and compared to the respective wild-type control. Error bars indicate the standard deviation for triplicate technical replicates. (E) Pol II occupancy at the CRC promoter determined by ChIP using anti-RPB2 antibodies in Ler and pwr-1. Pol II C1, located in the intergenic region between At2g17470 and At2g17460, has no appreciable Pol II occupancy as determined in a previous study [51] and is used as a negative control. (F) Col (left) and pwr-2 (right) plants grown side by side under 24-hour light conditions. RT-PCR analysis for PWR in Ler wild-type roots, rosette leaves, cauline leaves, and inflorescence tissues revealed high PWR transcript abundance in inflorescences (Figure 2C). To help establish a general idea of how PWR might function in various developmental pathways, microarray analysis was performed using pwr-2 and Col inflorescence tissue. Increased transcript levels of several genes with related functions in the specification of the floral meristem, including FLOWERING LOCUS T (FT), SUPPRESSOR OF CONSTANS 1 (SOC1), CAULIFLOWER (CAL), and SEPALLATA4 (SEP4), were observed in pwr-2 (fold-change ≥1.5, p-value≤0.005) (Table S2). FT acts upstream of SOC1, and both integrate signals from various flowering time pathways to induce flowering by promoting the expression of floral meristem identity genes including CAL and SEP4 [27]–[29]. Because these genes have closely related functions, it is likely that the transcript level changes observed for SOC1, CAL, and SEP4 were indirect consequences of an increase in FT or another more upstream factor. The increase in FT was confirmed using real-time RT-PCR for both pwr alleles (approximately 3 - to 4-fold increases, Figure 2D) and may be the underlying cause of the early flowering phenotype of the pwr mutants (Figure 2F).
Another related group of genes with altered transcript levels in pwr-2 were three direct targets of the floral homeotic genes APETALA3 (AP3) and PISTILLATA (PI): BANQUO1 (BNQ1) and BNQ2 and NAC-LIKE, ACTIVATED BY AP3/PI (NAP) [30], [31] (Table S1, Table S2). Although the changes in the transcript levels of all three genes suggested a decrease in AP3 and/or PI function, neither of them was identified as having a significant change in expression in the microarray analysis. Real-time RT-PCR analysis for PI and AP3 in pwr-2 revealed no significant change in PI transcript levels and a small reduction for AP3 (Figure 2D), which may explain the altered transcript levels of the AP3/PI target genes identified from the microarray analysis. However, the cause of the transcript level changes of AP3/PI targets remains unclear. Considering the role of AP3 and PI in specifying the second and third whorl floral organs (petals and stamens, respectively), these transcript level changes may provide a starting point for investigating the petal defects observed in pwr mutants.
PWR acts through AP2, CRC, and WUS in the floral determinacy network
To address whether PWR acts in any of the known pathways regulating floral determinacy, genetic analyses were carried out by combining pwr-1 and pwr-1 ag-10 with loss-of-function mutations in CLAVATA3 (CLV3), SUPERMAN (SUP), WUS, AG, CRC, and AP2.
Enhanced determinacy defects were found when clv3-1 was combined with ag-10 pwr-1. clv3-1 single mutant flowers produce slightly larger numbers of each floral organ type [32] including carpels (Figure 3A, Table 1). clv3-1 ag-10 flowers resembled clv3-1 flowers (Table 1). clv3-1 ag-10 pwr-1 triple mutant flowers were more indeterminate in several respects (Figure 3B). First, clv3-1 ag-10 pwr-1 produced a significantly larger number of carpels than clv3-1 and clv3-1 ag-10 (Table 1). Second, 47% of all triple mutant gynoecia that were dissected contained internal floral organs, which were never observed in clv3-1 or clv3-1 ag-10 (Table 1). The enhanced determinacy defects observed when ag-10 pwr-1 was combined with clv3-1 indicate that PWR confers determinacy in a pathway parallel to CLV3.
Fig. 3. Phenotypes of ag-10 pwr-1 in combination with loss-of-function mutations in other floral determinacy regulators. 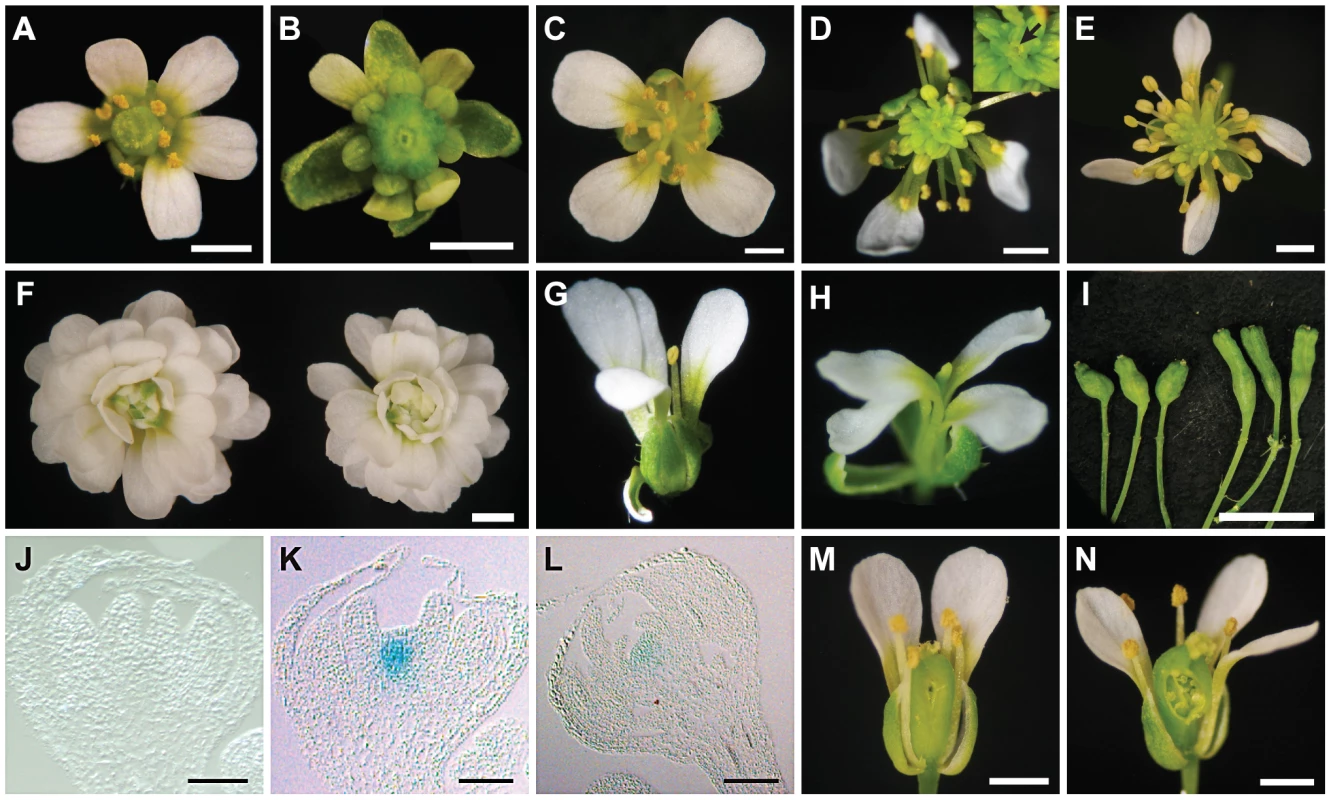
(A) clv3-1 flower with supernumerary organs in all four whorls. (B) clv3-1 ag-10 pwr-1 triple mutant flower. Compared to clv3-1, flowers of the triple mutant are small, have more carpels, and have shortened and bulged gynoecia often containing internal flowers. (C) sup-1 flower with aberrant carpels and more stamens than wild-type. (D) sup-1 ag-10 double mutant flower. These flowers are more strongly indeterminate compared to sup-1: more stamens are produced, and a mass of undifferentiated cells is visible at the center of the flower (magnified and indicated with an arrow in the inset). (E) sup-1 ag-10 pwr-1 triple mutant flower. (F) ag-1 (left) and ag-1 pwr-1 (right) flowers. pwr-1 does not enhance the indeterminacy defects of ag-1 flowers. (G) wus-1 flower exhibiting premature termination of floral development. (H) wus-1 ag-10 pwr-1 flower. Along with wus-1 pwr-1 (not shown), flowers of the triple mutant resemble wus-1 single mutant flowers. (I) ag-10 pwr-1 (left) and ap2-2 ag-10 pwr-1 (right) gynoecia. The ap2-2 mutation reduced the indeterminacy defects of ag-10 pwr-1: the triple mutant gynoecia were less bulged compared to ag-10 pwr-1, and a smaller percentage contained internal floral organs. (J) Longitudinal section of a stage 7 ag-10 pWUS:GUS flower showing the absence of GUS staining in the floral meristem. (K) Longitudinal section of a stage 7 or stage 8 ag-10 pwr-1 pWUS:GUS flower showing GUS staining in the floral meristem. (L) Longitudinal section of a stage 11 ag-10 pwr-1 pWUS:GUS flower. (M) crc-1 flower and (N) crc-1 pwr-1 flower with floral organs partially removed to reveal the gynoecia. While crc-1 single mutant flowers have unfused carpels and rarely exhibit indeterminacy defects, approximately half of all crc-1 pwr-1 gynoecia dissected contained internal floral organs. Scale bars = 10 µm in (J) to (L), 5 mm in (I), and 1 mm in all other panels. sup-1 mutant flowers produce supernumerary stamens, and the increased number of floral organs produced indicates a role for SUP as a positive regulator of floral determinacy [33]. sup-1 pwr-1 flowers had a small but statistically significant increase in organ number in the inner two whorls relative to sup-1 flowers (Table 1). ag-10 significantly enhanced the determinacy defects of sup-1 (Table 1): sup-1 ag-10 plants produced flowers with a significantly increased number of stamens and a visible group of undifferentiated cells in the center of the floral meristem (Figure 3D, Table 1). The sup-1 ag-10 pwr-1 triple mutant produced flowers similar to those of the sup-1 ag-10 double mutant (Figure 3E, Table 1). The lack of strong additive effects between pwr-1 and sup-1 suggests that PWR and SUP act in a common pathway in floral determinacy. However, how SUP impacts floral determinacy at the molecular level is unknown.
When crossed to ag-1, pwr-1 failed to enhance the determinacy defects of the null ag allele, based on flower size and the number of petals produced in ag-1 pwr-1 compared to ag-1 (Figure 3F). This suggested that PWR largely acted through the AG pathway to confer floral determinacy. Since AG is required for shutting off WUS expression in floral meristems at stage 6, further analysis was performed to assess whether PWR acted upstream of WUS in the regulation of floral determinacy. wus-1 pwr-1 and wus-1 ag-10 pwr-1 plants produced prematurely terminated wus-1-like flowers and failed to form the innermost floral organs (Figure 3G, 3H, wus-1 pwr-1 not shown). In addition to this analysis, crosses to a pWUS:GUS reporter line (for β-glucuronidase) containing a 3.2-kb WUS promoter [3], [34] were performed. In wild-type floral meristems, this reporter is active between floral stages 1 and 6 [3]. In the ag-10 background, GUS expression was only observed in approximately 10% of flowers beyond stage 6 (Figure 3J) [3], [34]. In ag-10 pwr-1, 90% of flowers (nine out of ten) showed prolonged GUS expression in late stage floral meristems (Figure 3K, 3L), indicating that WUS expression was prolonged in ag-10 pwr-1. Taken together, these analyses indicate that PWR confers determinacy in the same pathway as AG and that PWR acts upstream of WUS.
The YABBY transcription factor CRC has repeatedly been shown to promote floral determinacy, most demonstrably when crc-1 is combined with mutations in other floral determinacy genes [4]–[6]. The microarray analysis in pwr-2 indicated that CRC transcript levels were reduced in the mutant, and the ∼2-fold reduction was confirmed in pwr-1 and pwr-2 by real-time RT-PCR analysis (Table S1, Figure 2D). The relationship between PWR and CRC with respect to floral determinacy was further assessed by generating the crc-1 pwr-1 double mutant. Although neither crc-1 nor pwr-1 produced internal floral organs within the gynoecium, internal floral organs were observed in 76% of the crc-1 pwr-1 gynoecia dissected (Figure 3M, 3N, Table 1). This double mutant phenotype together with the reduced CRC transcript levels in both pwr mutants raises the possibility that PWR promotes floral determinacy through CRC and through a pathway parallel to CRC.
In contrast to AG, SUP, and CRC, which promote determinacy, AP2 promotes stem cell maintenance. AP2 has an antagonistic or complementary relationship with AG in the flower [8], [21]. Its role in floral stem cell maintenance has been evidenced by the severely indeterminate flowers produced when AP2 is simultaneously misexpressed and resistant to regulation by miR172 [8]. Compared to ag-10 pwr-1, ap2-2 ag-10 pwr-1 triple mutant plants exhibited a less severe indeterminate phenotype, indicated by reduced bulging and a reduced percentage of flowers containing internal floral organs (Figure 3I, Table 1). This suggested that the floral determinacy defects of ag-10 pwr-1 were partially due to increased AP2 expression or activity.
PWR promotes the expression of CRC and several MIR172 genes at the transcriptional level
Because AP2 is under the regulation of miR172 in the flower, experiments were performed to assess whether miR172 accumulation or activity was compromised by mutations in PWR. Small RNA northern blotting analysis was performed to measure the levels of miR172 in pwr-1 and pwr-2. The relative levels of miR172 in pwr-1 and pwr-2 were 0.6 and 0.3, respectively, compared to their corresponding wild-type controls (Figure 4A). Next, the levels of AP2 mRNA and AP2 protein in inflorescences were determined using real-time RT-PCR and western blotting. Differences in AP2 mRNA and protein levels were not observed between pwr-1 and Ler or between ag-10 pwr-1 and ag-10. This was not surprising because the expression of AP2 in the outer two whorls may have masked the effects of miR172 in the small number of floral stem cells. Utilizing a different approach, the 3′ cleavage products from the AP2 mRNA were detected to directly evaluate the activity of miR172. The levels of the 3′ cleavage products were reduced in pwr-1 relative to Ler and in ag-10 pwr-1 relative to ag-10 (Figure S1). Therefore, the reduced miR172 levels in pwr likely compromised the full repression of AP2 by miR172.
Fig. 4. miRNA abundance in pwr-1 and pwr-2. 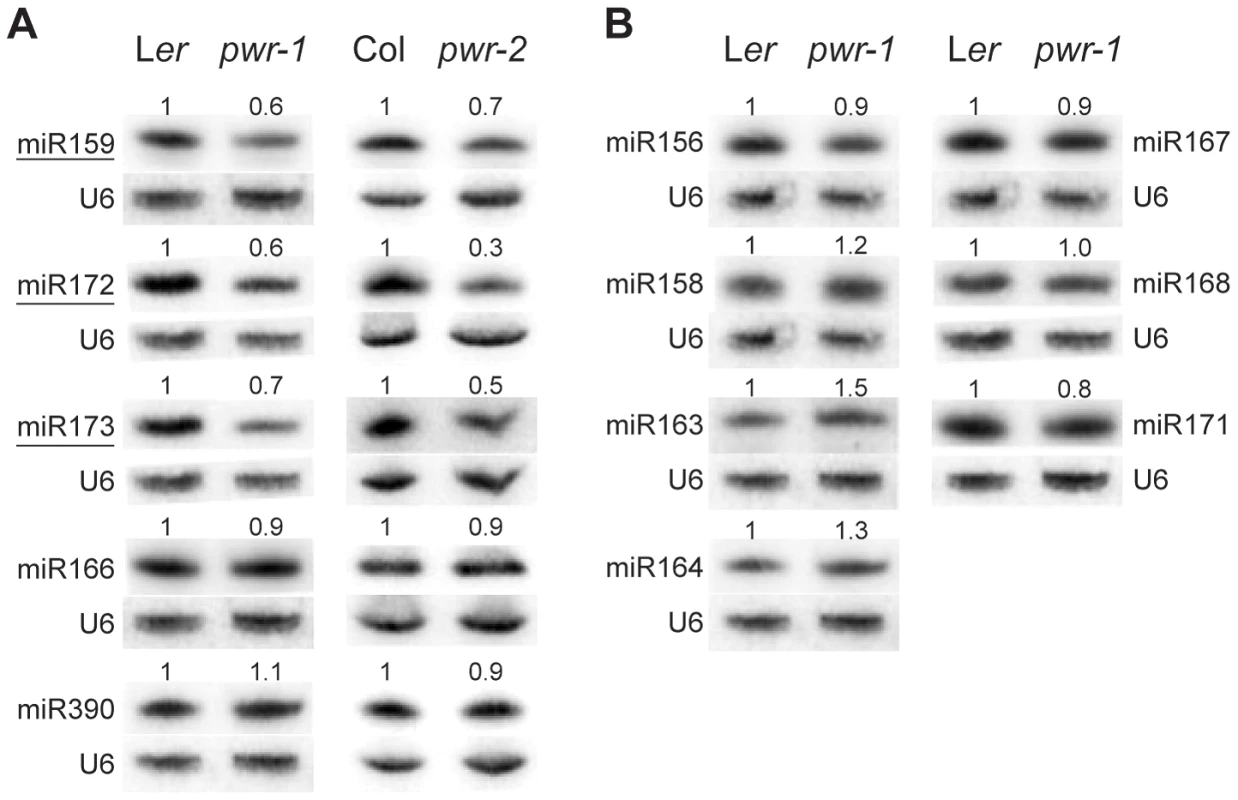
(A) Abundance of miR159, miR172, miR173, miR166, and miR390 in Ler, pwr-1, Col, and pwr-2 detected by small RNA northern blotting. The abundance of miR159, miR172, and miR173 (underlined) was reduced in both pwr alleles relative to their respective controls. Decreased accumulation was not observed for miR166 or miR390 in either allele. (B) Additional miRNAs tested in pwr-1 did not have reduced abundance compared to Ler. In (A) and (B), values indicate the relative abundance of the indicated miRNA species in pwr-1 compared to Ler or in pwr-2 compared to Col. For all blots, ImageJ signal intensity analysis was used for quantification. The numbers above the miRNA blots indicate the relative miRNA abundance between the mutants and wild type. The next question addressed was how PWR promotes the accumulation of miR172. Several lines of evidence suggested it was unlikely that miRNA biogenesis genes were compromised in the pwr mutants. First, northern blot analysis revealed that reductions in mature miRNA abundance were restricted to three (miR172, miR173, and miR159) of the examined miRNA species in both pwr alleles (Figure 4). Second, the expression levels of known miRNA biogenesis genes were not affected in pwr-2 according to the results from the microarray analysis. Additionally, there were no differences in pri-miR172a levels in PWR+ (both PWR/PWR and PWR/pwr) and pwr-1 plants harboring a p35S:MIR172a transgene (Figure 5B). Taken together, these findings suggested that the involvement of PWR in miRNA biogenesis is not downstream of transcription. To determine if PWR affected the transcription of individual MIR172 genes, real-time RT-PCR was performed for all five miR172 pri-miRNAs in pwr-1. Decreased transcript levels of pri-miR172a and pri-miR172b were observed in pwr-1, while the three remaining pri-miRNAs exhibited only subtle changes or were not affected (Figure 5A).
Fig. 5. pri–miRNA abundance and Pol II occupancy at MIR genes in pwr-1 and pwr-2. 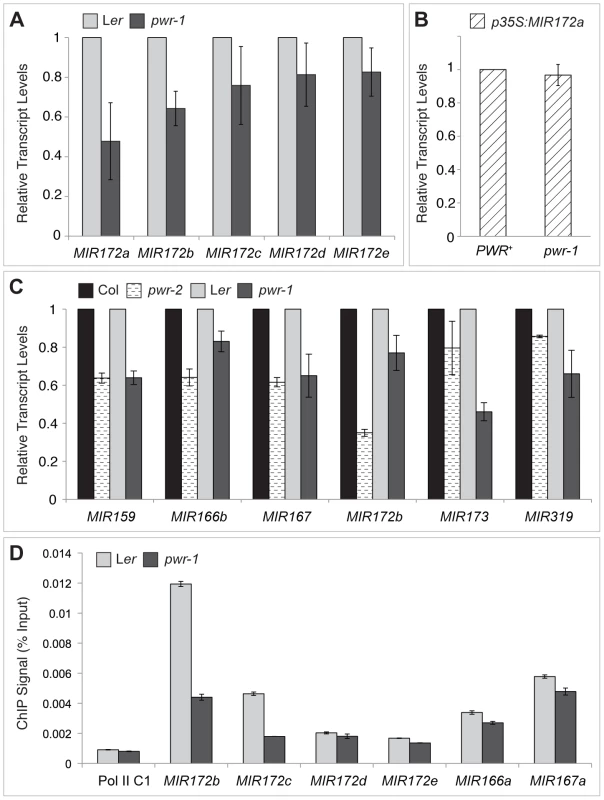
(A) Transcript levels of pri-miR172a-e in pwr-1. pri-miR172a and pri-miR172b transcript levels were reduced in pwr-1, while no significant changes were observed for pri-miR172c, pri-miR172d, and pri-miR172e. (B) Transcript levels of pri-miR172a in PWR+ (PWR/PWR and PWR/pwr-1) and pwr-1 plants harboring a p35S:MIR172a transgene. (C) Real-time RT-PCR analysis of other pri-miRNAs. A general decrease was observed in pri-miRNA transcript levels in both pwr alleles. For all pri-miRNAs tested, transcript levels were normalized to UBIQUITIN 5 and compared to the respective wild-type control. In (A) to (C), error bars indicate the standard deviation for triplicate technical replicates. Three biological replicates gave similar results. (D) Pol II occupancy at MIR loci determined by ChIP using anti-RPB2 antibodies in Ler and pwr-1. Error bars indicate the standard deviation for triplicate technical replicates. Pol II C1, located in the intergenic region between At2g17470 and At2g17460, has no appreciable Pol II occupancy as determined in a previous study [51] and is used as a negative control. To address the possibility that PWR might affect the transcription of other MIR genes, the abundance of other pri-miRNAs were determined using real-time RT-PCR. In both pwr-1 and pwr-2, compared to their respective wild-type controls, there was a general reduction in the transcript levels of the pri-miRNAs tested (pri-miR159, pri-miR166b, pri-miR167, pri-miR173, and pri-miR319) (Figure 5C). The occupancy of Pol II at the promoters of several miRNA genes and CRC was examined by chromatin immunoprecipitation (ChIP). Pol II occupancy was clearly reduced at the MIR172b, MIR172c, and CRC loci in pwr-1 (Figure 5D, Figure 2E), while only mild changes or no changes were observed at the MIR172d, MIR172e, MIR166a, and MIR167a loci (Figure 5D). These findings indicate that PWR specifically regulates CRC and some, but not all, of the MIR172 genes through the recruitment of Pol II to these loci.
MIR172d is required for floral determinacy
Coincidental with the observation that miR172 levels were reduced in pwr mutants, the mutation in another enhancer isolated from the ag-10 genetic screen was mapped to the MIR172d locus. A G-to-A mutation was identified at the ninth position of the 21-nucleotide sequence that corresponds to the mature miRNA (Figure 6D), and the mutation was designated mir172d-1. The enhanced floral determinacy phenotype of ag-10 mir172d-1 ranged from bulged gynoecia to the flower-within-flower phenotype, in which the fourth whorl organs were replaced with another flower (Figure 6A). The gynoecia of mir172d-1 single mutant flowers were not bulged but were shorter than those of Ler and occasionally consisted of three fused carpels instead of two (Figure 6B).
Fig. 6. Phenotype of the mir172d-1 single mutant and of mir172d-1 in combination with ag-10 and ap2-2. 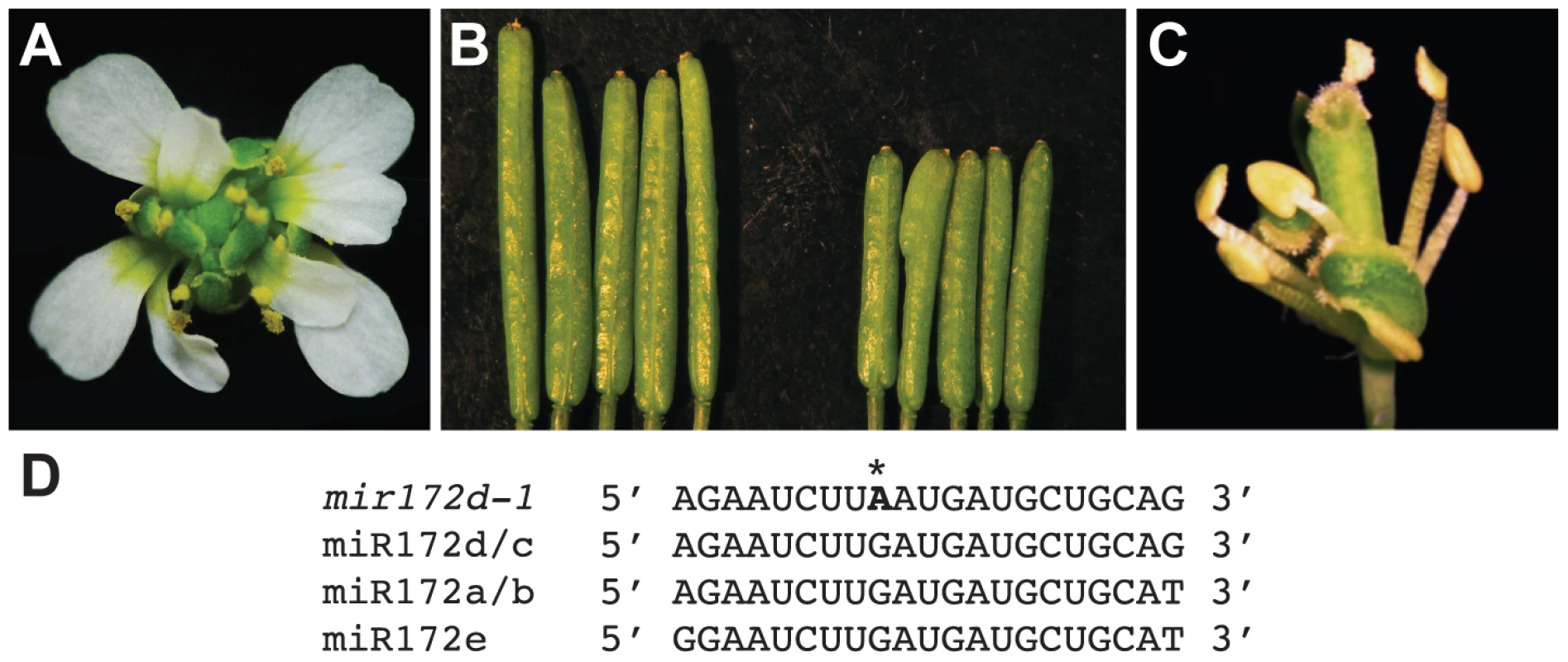
(A) ag-10 mir172d-1 flower. (B) Ler (left) and mir172d-1 (right) siliques. The gynoecia of mir172d-1 occasionally have three fused carpels instead of two. (C) ap2-2 ag-10 mir172d-1 flower. (D) Mature miR172 sequences and the site of the G-to-A mutation in mir172d-1 (indicated by *). To verify that MIR172d confers floral determinacy through the AP2 pathway, the status of miR172-mediated cleavage of AP2 mRNA was first evaluated in ag-10 and ag-10 mir172d-1 inflorescences. Semi-quantitative 5′ RACE PCR showed that the 3′ cleavage products from AP2 mRNA were reduced in ag-10 mir172d-1 relative to ag-10 (Figure S1), suggesting that miR172-mediated repression of AP2 was compromised. In addition, the ag-10 ap2-2 mir172d-1 triple mutant was generated to determine whether the floral determinacy defects of ag-10 mir172d-1 could be suppressed by ap2-2. The phenotype of the triple mutant clearly demonstrates that miR172-mediated repression of AP2 is required for floral determinacy as the ap2-2 mutation completely rescued the floral determinacy defects of ag-10 mir172d-1 (Figure 6C). Neither the flower-within-flower phenotype nor bulged carpels were observed in the triple mutant. Although a role for miR172 as a positive regulator of floral determinacy has previously been inferred from findings that established it as a negative regulator of AP2 in the flower, the ag-10 mir172d-1 phenotype provides direct loss-of-function evidence that at least one of the MIR172 genes, MIR172d, is required for the proper termination of the floral stem cells.
miR172 is also known to promote flowering by targeting several members of the AP2 gene family in addition to AP2 [18]. Interestingly, the mir172d-1 mutation did not affect flowering time (data not shown), suggesting that other MIR172 genes are sufficient to confer this developmental function.
Discussion
PWR contributes to the robustness of the floral determinacy network through MIR172 and CRC
The termination of the floral meristem requires various regulatory factors whose functions are orchestrated during the different stages of floral development. The data presented here indicate that a previously uncharacterized gene, PWR, confers floral determinacy through two distinct players: miR172 and CRC (Figure 7), which are active in the flower at different developmental stages. miR172 is present in the inner whorls from early stages onward [9], [21], while CRC is not expressed until carpel growth is initiated in late-stage flowers [35]. In addition to the possibility that the effect of PWR on floral stem cell termination is temporally distributed, the changes in miR172 accumulation and pri-miR172 transcript levels in pwr-1 shed light on how PWR contributes to the robustness of the floral determinacy network. The impact of PWR on multiple but not all of the five MIR172 genes and the finding that a mutation in MIR172d, whose expression is independent of PWR (Figure 7), also affects determinacy demonstrate that the expression diversification of this miRNA family is functionally relevant. Diversification in the regulation of the MIR172 genes and their common function in negatively regulating AP2 make the stem cell termination network more robust. This is reflected by the fact that loss of function in PWR (and consequently reduced expression of MIR172a-c) or MIR172d each has little effect on proper floral determinacy in the wild-type background.
Fig. 7. A summary of the floral determinacy gene network highlighting the function of PWR. 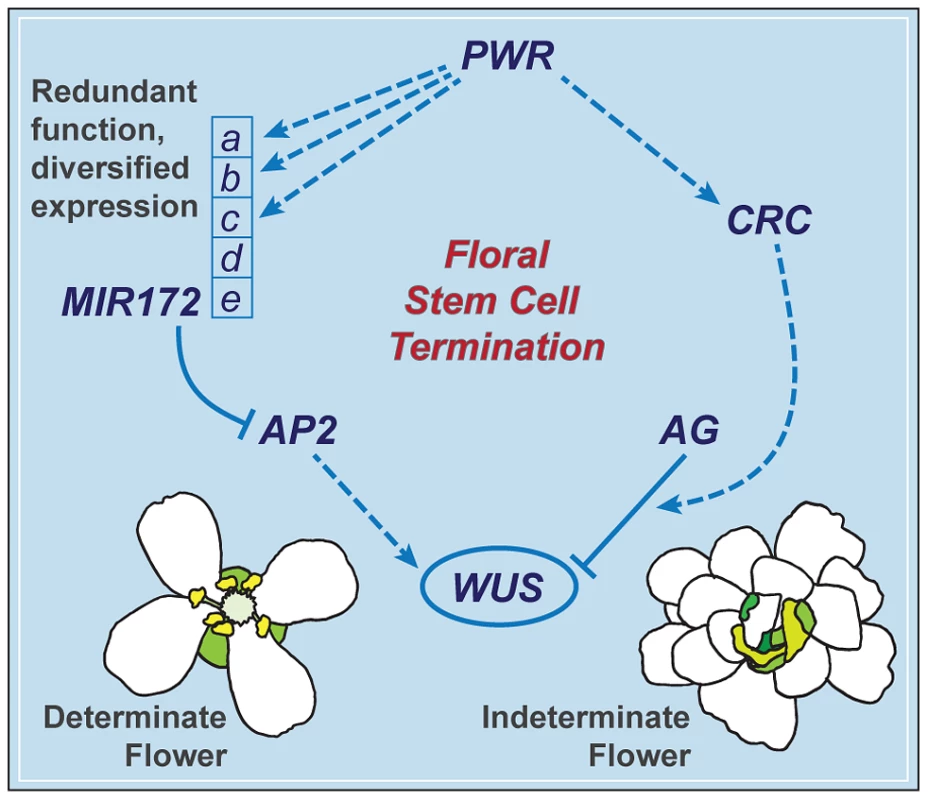
AP2 and AG act antagonistically in terms of the regulation of WUS expression: AP2 promotes WUS expression and stem cell maintenance, while AG represses WUS expression to elicit stem cell termination. CRC acts downstream of AG, and miR172 represses AP2 expression. PWR promotes floral stem cell termination by enhancing the expression of CRC and three of the five MIR172 genes. MIR172d, whose expression is independent of PWR, is functional in the repression of AP2 expression and the control of floral determinacy. PWR contributes to other floral developmental pathways
Although determinacy defects were not observed in single pwr mutants, the other pleiotropic defects initially observed in pwr-1 ag-10, including early flowering, petal defects, and small plant size, were observed in pwr-1. Some preliminary hypotheses can be drawn from the microarray and transcript level analysis, including the possible role of increased FT expression and decreased AP3 expression in disrupted flowering time and petal development, respectively. Although further investigation of the pleiotropic defects of pwr is required, the data clearly indicate that the defects are not attributable to widespread changes in miRNA abundance, as is the case in miRNA biogenesis mutants. First, the early flowering phenotype of pwr is in contrast to the late-flowering phenotype expected for a reduction of miR172 and is instead consistent with the increased FT transcript levels observed. Second, the widespread reduction observed for pri-miRNA levels in pwr were not accompanied by widespread decreases in mature miRNA abundance. One possible explanation for this discrepancy is that decreases in precursor levels do not always correspond to decreases in mature miRNAs, due in part to differences in the post-transcriptional maturation mechanisms for individual miRNAs [36], [37].
The predominant feature of the predicted PWR protein is a pair of SANT domains encoded in the fourth and largest exon of the gene. A BLAST search for proteins similar to PWR in Arabidopsis and other organisms identified PWR orthologs in other plant species but no homologs in Arabidopsis. Furthermore, PWR was found to have higher amino acid sequence similarity to histone deacetylase (HDAC) complex proteins in animals than to any other proteins in Arabidopsis. Studies in animals and yeast have uncovered the involvement of SANT domain-containing enzyme subunits in chromatin remodeling activity, including histone acetylation, histone deacetylation, and ATP-dependent chromatin remodeling [25]. The presence of two SANT domains in PWR and the spacing between them (173 amino acids) are similar to the domain structures of the HDAC subunits SMRT and N-CoR (nuclear receptor co-repressors), whose first and second SANT domains are necessary for HDAC activation and binding to unacetylated histone tails, respectively [38], [39]. Although further studies are necessary to determine whether PWR acts as a chromatin remodeling factor, this potential mode of activity may help explain how altered PWR function yields pleiotropic defects and changes in the transcript levels of otherwise unrelated genes, including MIR172B, CRC, and FT.
Individual MIR172 genes may have distinct developmental functions
In addition to its role in floral development, miR172 regulates flowering time as a negative regulator of floral repressors that belong to the AP2 transcription factor family [18]. The first characterization of miR172 function was made possible by transgenic lines in which miR172 was overexpressed (yielding early-flowering and ap2-like phenotypes) or lines in which the miR172 target AP2 was made resistant to regulation by the miRNA [9], [40]. Although the latter effectively corresponds to loss of miR172-mediated repression of AP2, it does not distinguish among the mature miR172 produced from the five MIR172 loci. Previous reports of differences in the accumulation of the individual pri-miRNAs have suggested variations in their individual functions throughout the plant. For example, the pri-miR172b precursor is particularly abundant in the shoot apex at the induction of flowering [41]. The miR172d-1 mutation provides direct genetic evidence for the role of miR172 as a positive regulator of floral determinacy, and the absence of flowering time defects in this mutant suggests that the developmental functions (flowering time and floral determinacy) assigned to miR172 can be uncoupled. Considered together with the effects of PWR on the transcription of some, but not all, of the MIR172 genes, these findings also demonstrate that the contributions of individual members of a miRNA family to one particular developmental process can be distinguished.
Materials and Methods
Plant materials and growth conditions
The mutants and transgenic lines used in this study are in the Landsberg erecta (Ler) ecotypic background, except for pwr-2, which is in the Columbia (Col) background, and ag-10Col, which is the ag-10 mutation introgressed into Col through six consecutive backcrosses. All plants were grown at 23°C under long-day conditions (16 hours light, 8 hours dark). ag-1 [23], clv3-1 [32], sup-1 [33], ap2-2 [42], wus-1 [43], crc-1 [4], and ag-10 [22] are previously described mutations.
pwr-1 and miR172d-1 were backcrossed into the ag-10 background and Ler two times prior to further analysis to reduce the number of extraneous background mutations. pwr-2 is a T-DNA insertion line, SALK_0713811C, obtained from the Arabidopsis Biological Resource Center [26].
Mapping of the pwr-1 and mir172d-1 mutations
The ag-10 pwr-1 and ag-10 mir172d-1 mutants were isolated from the ag-10 screen and crossed to ag-10Col to generate the respective mapping populations. For ag-10 pwr-1, 20 F2 plants exhibiting the enhanced phenotype were used for rough mapping, and linkage was observed for the ciw4 marker on the lower arm of chromosome 3. The mapping population was expanded to 888 plants for fine mapping. SSLP and CAPS markers were designed based on identified polymorphisms between the Ler and Col ecotypes (http://arabidopsis.org/browse/Cereon/index.jsp). The mapping region was narrowed to a 154-kb window encompassing portions of two BAC clones, F4F15 and T25B15, and spanning 55 open reading frames. A G-to-A mutation that produces a premature stop codon in At3g52250/PWR was identified by sequencing analysis. For ag-10 mir172d-1, 24 F2 plants exhibiting the enhanced phenotype were used for rough mapping, and linkage was again observed for the ciw4 marker on chromosome 3. Using an expanded mapping population of approximately 200 plants, the mapping region was narrowed to an 1,100-kb window spanning portions of the BAC clones T26I12 and T16L24. The sequencing analysis revealed a G-to-A mutation in At3g55512/MIR172d.
Genotyping
The primer sequences used for genotyping are listed in Table S3. For pwr-1, the dCAPS primers PWRgeno-F and PWRgeno-R produce an HphI (NEB, Cat# R0158) site exclusively in the mutant. The pwr-2 mutation was genotyped using primers EN1_XcmIF and 3G52250R1, which fail to amplify a fragment in the homozygous mutant. The ag-10 mutation eliminates an HpyAV restriction site and was genotyped using primers AGp1 and ag10_genoR followed by HpyAV digestion (NEB, Cat# R0621). For miR172d-1, the primers R194geno-F and R194geno-R were used to amplify the genomic fragment. The fragment amplified from miR172d-1 fails to be cut by Hpy188III (NEB, Cat# R0622).
Plasmid construction
To generate pPWR:PWR-GFP, the PWR genomic region was amplified using primers EN1_full_CACC and EN1cDNA_NS (Table S3). The 8.1-kb genomic region includes 1,670 bp of the upstream promoter region and the entire coding region of PWR, excluding the stop codon and three preceding nucleotides. The fragment was cloned into pENTR/D-TOPO (Invitrogen, Cat# K2400-20) and introduced into the pMDC107 Gateway-compatible vector [44]. ag-10 pwr-1 and pwr-1 plants were transformed with the plasmid by agroinfiltration for the phenotypic rescue analysis.
RNA extraction and real-time RT–PCR
The harvesting of inflorescence tissue was consistently performed at the same time of day. RNA was extracted using TRI reagent (MRC, Cat# TR118), and DNA was removed using DNase I (Roche, Cat# 04716728001). Oligo-dT primer (Fermentas, Cat# S0131) and reverse transcriptase (Fermentas, Cat# EP0441) were used to synthesize cDNA. All procedures used were according to the manufacturers' instructions. Quantitative PCR was performed in triplicate on a Bio-Rad IQ cycler apparatus using iQ SYBR Green Supermix (Bio-Rad, Cat#170-0082). The primers used for real-time RT-PCR were previously described or designed using the Primer3 online tool (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) and are listed in Table S3.
Small RNA Northern blot analysis
The hybridization and detection of miRNAs were performed as previously described [45]. Five micrograms total RNA was obtained from inflorescence tissues, and 5′-end-labeled 32P antisense DNA oligonucleotides were used to detect mature miRNA species. U6 was used as the loading control. The oligonucleotide probes used were previously described [46], [47] and are listed in Table S3. The signal intensities of the blots were quantified using ImageJ processing and analysis software (http://rsbweb.nih.gov/ij/).
ATH1 Affymetrix microarray analysis
Total RNA extracted from inflorescences was purified using the RNeasy Plant Mini Kit (Qiagen, Cat# 74903). Affymetrix Gene Chip probe preparation, hybridization, and quality control were performed according to the manufacturer's instructions. The SimpleAffy package available for the R language was used to analyze the microarray data, and expression values were determined using the MAS 5.0 method as previously described [48]. Changes in gene expression were defined as transcript level fold-changes ≥1.5 with p-values≤0.005.
Histochemical staining
GUS staining was performed as previously described [49], [50]. Inflorescences were fixed in 90% cold acetone for 15 to 20 min and rinsed with the rinse solution [50 mM NaPO4, pH 7.2; 0.5 mM K3Fe(CN)6; and 0.5 mM K4Fe(CN)6]. After the infiltration solution [50 mM NaPO4, pH 7.2; 0.5 mM K3Fe(CN)6; 0.5 mM K4Fe(CN)6; and 2 mM X-Gluc] was added, the inflorescences were vacuum infiltrated for 10 min then incubated at 37°C overnight.
Chromatin immunoprecipitation
ChIP was performed as previously described [3]. Inflorescences were ground in liquid nitrogen and cross-linked with 1% formaldehyde in M1 buffer [10 mM phosphate buffer, pH 7.0; 0.1 M NaCl; 10 mM mercaptoethanol; 1 M hexylene glycol, 1× protease inhibitor cocktail (Roche); and 1 mM PMSF] for 10 min. The suspension was filtered through four layers of Miracloth, and the filtrate was centrifuged at 12,000 rpm for 10 min. The pelleted chromatin was washed three times with M2 buffer (M1 buffer plus 10 mM MgCl2 and 0.5% Triton X-100) and one time with M3 buffer [10 mM phosphate buffer, pH 7.0; 0.1 M NaCl; 10 mM mercaptoethanol; 1× protease inhibitor cocktail (Roche); and 1 mM PMSF]. The chromatin was resuspended in nuclei lysis buffer and sonicated to generate DNA fragments approximately 500 bp in length. The lysate was pre-cleared by incubation with 50 µL protein-A agarose beads/salmon sperm DNA (Millipore) for 1 h then incubated with anti-RBP2 antibody (Abcam 10338) overnight. The bound DNA fragments were recovered and purified using columns from the Qiagen Plasmid Extraction Kit according to the manufacturer's instructions. Quantitative real-time PCR was performed on bound and input DNAs. The primers used are listed in Table S3.
5′ RACE to detect 3′ cleavage products from AP2 mRNA
Total RNA was extracted from dissected inflorescences, and mRNA was isolated using the Sera-Mag Magnetic Oligo(dT) kit (Thermo Scientific) according to the manufacturer's instructions. 5′ RACE was performed using the GeneRacer Kit (Invitrogen) according to the manufacturer's instructions. Briefly, an RNA oligonucleotide adaptor was ligated to the isolated mRNAs, and reverse transcription was conducted using oligo-dT. AP2 3′ cleavage products were amplified and sequenced. For amplification, a primer specific to the 5′ adaptor and an AP2-specific primer were used.
Supporting Information
Zdroje
1. LenhardM, BohnertA, JürgensG, LauxT (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105 : 805–814.
2. SunB, XuY, NgKH, ItoT (2009) A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev 23 : 1791–1804.
3. LiuX, KimYJ, MüllerR, YumulRE, LiuC, et al. (2011) AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23 : 3654–3670.
4. AlvarezJ, SmythDR (1999) CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126 : 2377–2386.
5. PrunetN, MorelP, ThierryAM, EshedY, BowmanJL, et al. (2008) REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell 20 : 901–919.
6. Zúñiga-MayoVM, Marsch-MartínezN, de FolterS (2012) JAIBA, a class-II HD-ZIP transcription factor involved in the regulation of meristematic activity, and important for correct gynoecium and fruit development in Arabidopsis. Plant J 71 : 314–326.
7. WürschumT, Gross-HardtR, LauxT (2006) APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell 18 : 295–307.
8. ZhaoL, KimY, DinhTT, ChenX (2007) miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J 51 : 840–849.
9. ChenX (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303 : 2022–2025.
10. VoinnetO (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136 : 669–687.
11. ChenX (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25 : 21–44.
12. Rubio-SomozaI, WeigelD (2011) MicroRNA networks and developmental plasticity in plants. Trends Plant Sci
13. LeeY, KimM, HanJ, YeomKH, LeeS, et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23 : 4051–4060.
14. XieZ, AllenE, FahlgrenN, CalamarA, GivanSA, et al. (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138 : 2145–2154.
15. FahlgrenN, JogdeoS, KasschauKD, SullivanCM, ChapmanEJ, et al. (2010) MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22 : 1074–1089.
16. CuperusJT, FahlgrenN, CarringtonJC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23 : 431–442.
17. MaherC, SteinL, WareD (2006) Evolution of Arabidopsis microRNA families through duplication events. Genome Res 16 : 510–519.
18. ZhuQH, HelliwellCA (2011) Regulation of flowering time and floral patterning by miR172. J Exp Bot 62 : 487–495.
19. RajagopalanR, VaucheretH, TrejoJ, BartelDP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20 : 3407–3425.
20. GrigorovaB, MaraC, HollenderC, SijacicP, ChenX, et al. (2011) LEUNIG and SEUSS co-repressors regulate miR172 expression in Arabidopsis flowers. Development 138 : 2451–2456.
21. WollmannH, MicaE, TodescoM, LongJA, WeigelD (2010) On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137 : 3633–3642.
22. JiL, LiuX, YanJ, WangW, YumulRE, et al. (2011) ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet 7: e1001358 doi:10.1371/journal.pgen.1001358.
23. BowmanJL, SmythDR, MeyerowitzEM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1 : 37–52.
24. NohYS, AmasinoRM (2003) PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15 : 1671–1682.
25. BoyerLA, LatekRR, PetersonCL (2004) The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol 5 : 158–163.
26. AlonsoJM, StepanovaAN, LeisseTJ, KimCJ, ChenH, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 : 653–657.
27. LiuC, ThongZ, YuH (2009) Coming into bloom: the specification of floral meristems. Development 136 : 3379–3391.
28. FornaraF, de MontaiguA, CouplandG (2010) SnapShot: Control of flowering in Arabidopsis. Cell 141 : 550, 550.e551–552.
29. DittaG, PinyopichA, RoblesP, PelazS, YanofskyMF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14 : 1935–1940.
30. SablowskiRW, MeyerowitzEM (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92 : 93–103.
31. MaraCD, HuangT, IrishVF (2010) The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell 22 : 690–702.
32. ClarkSM, RunningMP, MeyerowitzEM (1995) CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121 : 2057–2067.
33. BowmanJL, SakaiH, JackT, WeigelD, MayerU, et al. (1992) SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114 : 599–615.
34. BäurleI, LauxT (2005) Regulation of WUSCHEL transcription in the stem cell niche of the Arabidopsis shoot meristem. Plant Cell 17 : 2271–2280.
35. BowmanJL, SmythDR (1999) CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126 : 2387–2396.
36. WinterJ, JungS, KellerS, GregoryRI, DiederichsS (2009) Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11 : 228–234.
37. VolkN, ShomronN (2011) Versatility of MicroRNA biogenesis. PLoS ONE 6: e19391 doi:10.1371/journal.pone.0019391.
38. GuentherMG, BarakO, LazarMA (2001) The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol 21 : 6091–6101.
39. YuJ, LiY, IshizukaT, GuentherMG, LazarMA (2003) A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J 22 : 3403–3410.
40. AukermanMJ, SakaiH (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15 : 2730–2741.
41. SchmidM, UhlenhautNH, GodardF, DemarM, BressanR, et al. (2003) Dissection of floral induction pathways using global expression analysis. Development 130 : 6001–6012.
42. BowmanJL, SmythDR, MeyerowitzEM (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112 : 1–20.
43. LauxT, MayerK, BergerJ, JurgensG (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122 : 87–96.
44. CurtisMD, GrossniklausU (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 : 462–469.
45. ParkW, LiJ, SongR, MessingJ, ChenX (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12 : 1484–1495.
46. YuB, BiL, ZhengB, JiL, ChevalierD, et al. (2008) The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci U S A 105 : 10073–10078.
47. KimYJ, ZhengB, YuY, WonSY, MoB, et al. (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30 : 814–822.
48. HoranK, JangC, Bailey-SerresJ, MittlerR, SheltonC, et al. (2008) Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol 147 : 41–57.
49. Rodrigues-PousadaRA, De RyckeR, DedonderA, Van CaeneghemW, EnglerG, et al. (1993) The Arabidopsis 1-Aminocyclopropane-1-Carboxylate Synthase Gene 1 Is Expressed during Early Development. Plant Cell 5 : 897–911.
50. JeffersonRA, KavanaghTA, BevanMW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 : 3901–3907.
51. ChodavarapuRK, FengS, BernatavichuteYV, ChenPY, StroudH, et al. (2010) Relationship between nucleosome positioning and DNA methylation. Nature 466 : 388–392.
Štítky
Genetika Reprodukční medicína
Článek Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across PathogensČlánek TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association StudiesČlánek Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization inČlánek Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA ExpressionČlánek The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of GenesČlánek The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 1
-
Všechny články tohoto čísla
- A Model of High Sugar Diet-Induced Cardiomyopathy
- Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across Pathogens
- Emerging Function of Fat Mass and Obesity-Associated Protein (Fto)
- Positional Cloning Reveals Strain-Dependent Expression of to Alter Susceptibility to Bleomycin-Induced Pulmonary Fibrosis in Mice
- Genetics of Ribosomal Proteins: “Curiouser and Curiouser”
- Transposable Elements Re-Wire and Fine-Tune the Transcriptome
- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- MAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
- Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies
- A Systematic Mapping Approach of 16q12.2/ and BMI in More Than 20,000 African Americans Narrows in on the Underlying Functional Variation: Results from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
- Histone H3K56 Acetylation, Rad52, and Non-DNA Repair Factors Control Double-Strand Break Repair Choice with the Sister Chromatid
- Genome-Wide Association Study Identifies a Novel Susceptibility Locus at 12q23.1 for Lung Squamous Cell Carcinoma in Han Chinese
- Genetic Disruption of the Copulatory Plug in Mice Leads to Severely Reduced Fertility
- The [] Prion Exists as a Dynamic Cloud of Variants
- Adult Onset Global Loss of the Gene Alters Body Composition and Metabolism in the Mouse
- Fis Protein Insulates the Gene from Uncontrolled Transcription
- The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse
- Genome-Wide Haplotype Analysis of Expression Quantitative Trait Loci in Monocytes
- TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association Studies
- Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
- The Ecm11-Gmc2 Complex Promotes Synaptonemal Complex Formation through Assembly of Transverse Filaments in Budding Yeast
- MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
- Comparative Genomic Analysis of the Endosymbionts of Herbivorous Insects Reveals Eco-Environmental Adaptations: Biotechnology Applications
- Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry
- Pharmacodynamics, Population Dynamics, and the Evolution of Persistence in
- A Hybrid Likelihood Model for Sequence-Based Disease Association Studies
- Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity
- Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis
- Transcriptional Dynamics Elicited by a Short Pulse of Notch Activation Involves Feed-Forward Regulation by Genes
- Dynamic Large-Scale Chromosomal Rearrangements Fuel Rapid Adaptation in Yeast Populations
- Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability
- Gene Copy-Number Polymorphism Caused by Retrotransposition in Humans
- An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in
- Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization in
- Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination
- Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes and
- Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene
- A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
- Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators
- Genome-Wide Analysis Reveals Selection for Important Traits in Domestic Horse Breeds
- Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
- Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
- HIF- and Non-HIF-Regulated Hypoxic Responses Require the Estrogen-Related Receptor in
- Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages
- The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Loci Associated with -Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers
- Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol
- Centromere-Like Regions in the Budding Yeast Genome
- Sequencing of Loci from the Elephant Shark Reveals a Family of Genes in Vertebrate Genomes, Forged by Ancient Duplications and Divergences
- Mendelian and Non-Mendelian Regulation of Gene Expression in Maize
- Mutational Spectrum Drives the Rise of Mutator Bacteria
- Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression
- The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of Genes
- Sex-Specific Signaling in the Blood–Brain Barrier Is Required for Male Courtship in
- A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity
- Is Required for Leptin-Mediated Depolarization of POMC Neurons in the Hypothalamic Arcuate Nucleus in Mice
- Unlocking the Bottleneck in Forward Genetics Using Whole-Genome Sequencing and Identity by Descent to Isolate Causative Mutations
- The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
- MTERF3 Regulates Mitochondrial Ribosome Biogenesis in Invertebrates and Mammals
- Downregulation and Altered Splicing by in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
- NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses
- Retroactive Maintains Cuticle Integrity by Promoting the Trafficking of Knickkopf into the Procuticle of
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network
- Genetic and Functional Modularity of Activities in the Specification of Limb-Innervating Motor Neurons
- A Population Genetic Model for the Maintenance of R2 Retrotransposons in rRNA Gene Loci
- A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in
- A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1)
- Mutation of the Diamond-Blackfan Anemia Gene in Mouse Results in Morphological and Neuroanatomical Phenotypes
- Life, the Universe, and Everything: An Interview with David Haussler
- Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome Oxidase Blockade and Limits Mitochondrial ROS Overproduction
- An Evolutionarily Conserved Synthetic Lethal Interaction Network Identifies FEN1 as a Broad-Spectrum Target for Anticancer Therapeutic Development
- The Flowering Repressor Underlies a Novel QTL Interacting with the Genetic Background
- Telomerase Is Required for Zebrafish Lifespan
- and Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
- WNK1/HSN2 Mutation in Human Peripheral Neuropathy Deregulates Expression and Posterior Lateral Line Development in Zebrafish ()
- Synergistic Interaction of Rnf8 and p53 in the Protection against Genomic Instability and Tumorigenesis
- Dot1-Dependent Histone H3K79 Methylation Promotes Activation of the Mek1 Meiotic Checkpoint Effector Kinase by Regulating the Hop1 Adaptor
- A Heterogeneous Mixture of F-Series Prostaglandins Promotes Sperm Guidance in the Reproductive Tract
- Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
- Directed Evolution of a Model Primordial Enzyme Provides Insights into the Development of the Genetic Code
- Genome-Wide Screens for Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



