-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaDNA Damage and Reactive Nitrogen Species are Barriers to Colonization of the Infant Mouse Intestine
Ingested Vibrio cholerae pass through the stomach and colonize the small intestines of its host. Here, we show that V. cholerae requires at least two types of DNA repair systems to efficiently compete for colonization of the infant mouse intestine. These results show that V. cholerae experiences increased DNA damage in the murine gastrointestinal tract. Agreeing with this, we show that passage through the murine gut increases the mutation frequency of V. cholerae compared to liquid culture passage. Our genetic analysis identifies known and novel defense enzymes required for detoxifying reactive nitrogen species (but not reactive oxygen species) that are also required for V. cholerae to efficiently colonize the infant mouse intestine, pointing to reactive nitrogen species as the potential cause of DNA damage. We demonstrate that potential reactive nitrogen species deleterious for V. cholerae are not generated by host inducible nitric oxide synthase (iNOS) activity and instead may be derived from acidified nitrite in the stomach. Agreeing with this hypothesis, we show that strains deficient in DNA repair or reactive nitrogen species defense that are defective in intestinal colonization have decreased growth or increased mutation frequency in acidified nitrite containing media. Moreover, we demonstrate that neutralizing stomach acid rescues the colonization defect of the DNA repair and reactive nitrogen species defense defective mutants suggesting a common defense pathway for these mutants.
Published in the journal: . PLoS Pathog 7(2): e32767. doi:10.1371/journal.ppat.1001295
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001295Summary
Ingested Vibrio cholerae pass through the stomach and colonize the small intestines of its host. Here, we show that V. cholerae requires at least two types of DNA repair systems to efficiently compete for colonization of the infant mouse intestine. These results show that V. cholerae experiences increased DNA damage in the murine gastrointestinal tract. Agreeing with this, we show that passage through the murine gut increases the mutation frequency of V. cholerae compared to liquid culture passage. Our genetic analysis identifies known and novel defense enzymes required for detoxifying reactive nitrogen species (but not reactive oxygen species) that are also required for V. cholerae to efficiently colonize the infant mouse intestine, pointing to reactive nitrogen species as the potential cause of DNA damage. We demonstrate that potential reactive nitrogen species deleterious for V. cholerae are not generated by host inducible nitric oxide synthase (iNOS) activity and instead may be derived from acidified nitrite in the stomach. Agreeing with this hypothesis, we show that strains deficient in DNA repair or reactive nitrogen species defense that are defective in intestinal colonization have decreased growth or increased mutation frequency in acidified nitrite containing media. Moreover, we demonstrate that neutralizing stomach acid rescues the colonization defect of the DNA repair and reactive nitrogen species defense defective mutants suggesting a common defense pathway for these mutants.
Introduction
Maintaining genomic integrity during infection is important for several bacterial pathogens to colonize their hosts. DNA repair defects in Listeria monocytogenes, Salmonella typhimurium, Helicobacter pylori and others leads to decreased or even a complete attenuation of virulence [1]–[6]. While there are several types of DNA repair in bacteria [7], many of the studies showing a requirement for DNA repair in pathogenesis focus on three pathways: the SOS response, base excision repair, and mismatch repair [1]–[10].
The SOS response is a well studied and conserved stress response in bacteria that is elicited following DNA damage and replication fork arrest [for review [11]]. The SOS response is controlled by positive and negative regulators. RecA positively regulates the SOS response by binding to single-stranded DNA fragments generated by attempted replication past DNA lesions. The RecA/ssDNA nucleoprotein filament induces the auto-cleavage of the negative regulator LexA, a transcriptional repressor. Cleavage of LexA allows for expression of 57 genes in the E. coli SOS regulon including translesion DNA polymerases that are able to replicate DNA past noncoding base lesion, and proteins involved in the inhibition of cell division [for review [11]].
Base excision repair (BER) is the most common form of repair for single base damage [for review [7]]. In BER, a DNA N-glycosylase first excises the damaged base from the deoxyribose moiety in the DNA strand creating an abasic site. Class II apurinic/apyrimidinic (AP) endonuclease then hydrolyzes the phosphodiester bond immediately 5′ to the abasic site [for review (Kornberg and Baker 1992, Friedberg 2005)]. Subsequent actions process this site to prime and repair the abasic site ultimately by DNA synthesis with DNA polymerase I and ligation by DNA ligase.
Normal replication can also introduce errors in the form of mismatched DNA base-pairs. These mismatches can lead to permanent mutations after a subsequent round of DNA replication. Mismatch repair (MMR) specifically identifies and corrects these base pairing errors increasing the fidelity of the replication pathway nearly ∼1000-fold [for review [7]].
While extensive work has shown the benefit of maintaining genomic integrity for an invading bacterium, there appear to be instances where lapses in genomic fidelity are beneficial for a pathogenic bacterium [12]–[14]. In order to colonize and thrive in a mammalian host, a bacterium must be able to adapt and respond to the conditions and stresses associated its new environment. Genomic mutations support this by allowing current gene products to gain or alter their functions. The utility of mutation(s) and a pathogen's ability to grow in the human environment has been a source of discussion for several years [13], [15]. Giraud et al. showed that a high mutation rate was initially beneficial for Escherichia coli to colonize the mouse gut, but this benefit became a liability once adaptation had been reached [12]. Oliver et al. demonstrated that Pseudomonas aeruginosa from chronically infected individuals often has an increased mutation frequency, suggesting an increased mutation rate can be beneficial to P. aeruginosa to allow rapid adaptation to the hostile host environment [14]. Thus depending on the pathogen, the mode and duration of the infection, defects in DNA repair may be detrimental or beneficial to the infecting bacterium.
Several studies have indicated that host produced reactive oxygen species (ROS) and reactive nitrogen species (RNS) cause DNA damage to the invading bacterium [4], [16], [17]. Not surprisingly bacteria have several defense mechanisms to detoxify ROS and RNS. Each enzyme detoxifies a specific type of ROS or RNS. For example catalases/peroxidases decompose H2O2, superoxide dismutases dismutate superoxide and ferrisiderophore reductase removes nitric oxide [18], [19]. As with certain DNA repair systems, loss of ROS and RNS defenses have been shown to attenuate bacterial pathogens [16], [20].
Studies supporting the importance of ROS/RNS defenses and DNA repair pathways in bacterial pathogenesis often focus on intracellular pathogens [1], [2], [4]. To survive, intracellular pathogens engulfed by phagocytic cells are either able to escape the phagosome or have mechanisms to survive within it. Within the phagosome, captured bacteria may be exposed to host production of ROS and RNS in a host defense response called the oxidative burst. It is hypothesized that the oxidative burst is responsible for the DNA damage experienced by engulfed bacteria [4], [16], [17].
Vibrio cholerae is the causative agent of the severe human diarrheal disease cholera. V. cholerae is a non-invasive pathogen that colonizes the small intestine of its host [21], [22]. As a non-invasive pathogen, V. cholerae is not expected to experience the same types of stresses as intracellular pathogens, such as an oxidative burst. However V. cholerae does pass through several hostile environments as the disease progresses. Immediately following ingestion, V. cholerae is exposed to the exceptionally antagonistic environment of the stomach where the pH of gastric acid can reach as low as 1 [23], [24]. Furthermore, nitrite from both food sources and the salivary nitrite cycle can enter the stomach creating acidified nitrite [25], [26], [27]. Acidified nitrite has potent antimicrobial effects on gut pathogens [28], [29], [30], [31]. These studies show that the viability of several pathogenic bacteria decreases rapidly under acidified nitrite conditions. Furthermore, nitrates, which can also be found in the stomach, have been shown to modify of gene expression reducing acid tolerance [32]. The antimicrobial effects of acidified nitrite are thought to be due to the generation of deleterious RNS [33]. However, with the exception of a few studies [34], [35], [36], the points of action of these RNS as well as the bacterial determinants required for protection against them have remained largely unexamined. After traversing the stomach V. cholerae faces several innate host defenses in the intestine including bile, lysozyme, small antimicrobial peptides and complement [37]. Thus, V. cholerae must overcome several barriers during infection that have the potential to cause DNA damage through a direct or indirect mechanism.
We report here that V. cholerae strain C6706 experiences increased DNA damage during passage through the murine gastrointestinal track. We demonstrate that increased genomic stress is a potential barrier to host colonization by V. cholerae. We found that two important DNA repair pathways are necessary for V. cholerae to efficiently colonize the infant mouse intestine. Furthermore, we show that defense against RNS is also necessary for V. cholerae to colonize the infant mouse. In doing so we identify a novel protein required for defense against RNS in pathogenic bacteria. In vitro we show that all our colonization defective DNA repair and RNS defense mutants share a common sensitivity to acidified nitrite and we further show that neutralizing stomach acid rescues intestinal colonization defect of these mutants.
Results
Mismatch repair and base excision repair pathways are required for V. cholerae colonization of the infant mouse intestine
To determine if V. cholerae requires defenses against DNA damage during colonization, we tested a series of transposon mutants that contained insertions in different steps in three important DNA repair pathways for their ability to colonize the infant mouse intestine in competition with the wild type strain. These pathways were nucleotide excision repair (NER), base excision repair (BER) and mismatch repair (MMR) (Table 1). While the SOS response is an important contributor to genomic integrity we did not test a requirement for SOS since Quiones et al. previously showed that SOS activation is not required for intestinal cholera toxin production or colonization [38]. We used uvrA as a representative gene required for NER since uvrA is obligatory for NER. We found no difference in the ability of the uvrA::Tn containing strain to colonize the infant mouse relative to the parental strain suggesting that NER is dispensable for V. cholerae pathogeneis (Table 1).
Tab. 1. Ability of V. cholerae mutants defective in DNA repair pathways to colonize the infant mouse intestine in competition with the parental strain (WT). 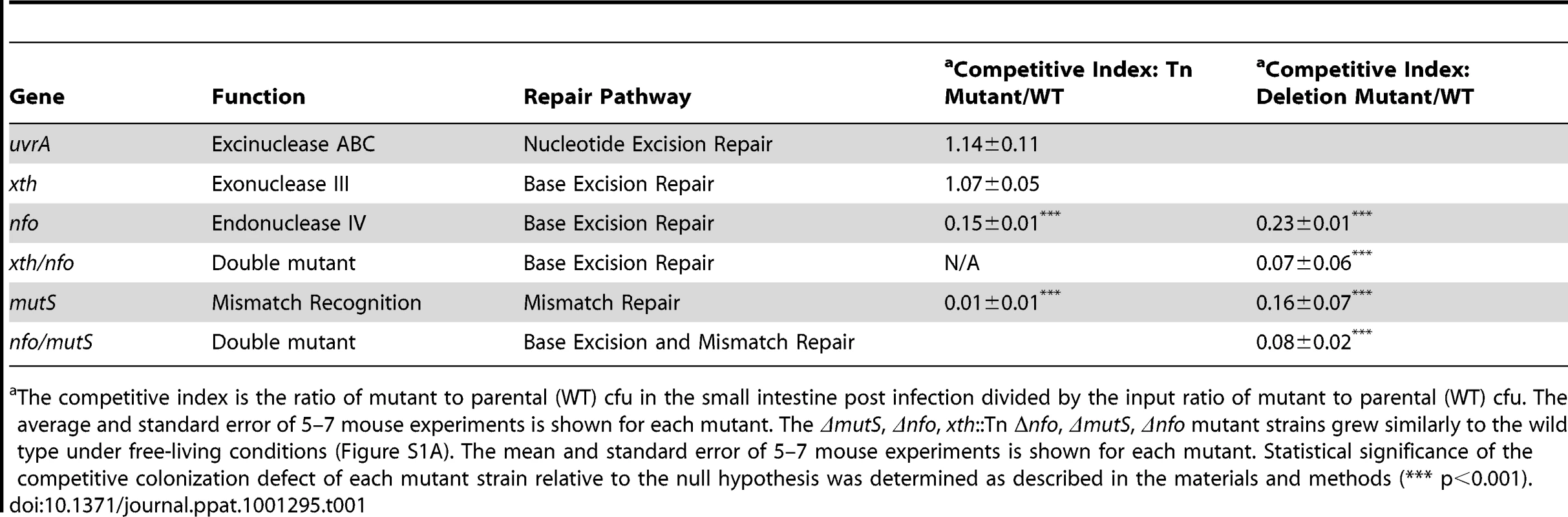
The competitive index is the ratio of mutant to parental (WT) cfu in the small intestine post infection divided by the input ratio of mutant to parental (WT) cfu. The average and standard error of 5–7 mouse experiments is shown for each mutant. The ΔmutS, Δnfo, xth::Tn Δnfo, ΔmutS, Δnfo mutant strains grew similarly to the wild type under free-living conditions (Figure S1A). The mean and standard error of 5–7 mouse experiments is shown for each mutant. Statistical significance of the competitive colonization defect of each mutant strain relative to the null hypothesis was determined as described in the materials and methods (*** p<0.001). Apurinic/apyrimidininc (AP) endonucleases are critical in BER. BER has been most well studied in E. coli. E. coli encodes two class II AP endonucleases, Xth [endo II (endo VI)] and Nfo (endo IV). In E. coli Xth is responsible for ∼90% of the AP endonuclease activity in the cell [39], [40]. Few phenotypes have been attributed solely to Nfo activity but Nfo is known to contribute to BER [41]. V. cholerae carries close homologs of both Xth and Nfo (VC1860 and VC2360 respectively). Interestingly we found that the xth::Tn mutant was not defective in intestinal colonization however the nfo::Tn mutant showed a defect in colonization compared to the parental strains (Table 1). We created a clean deletion of nfo (Δnfo) in V. cholerae and found this mutant also had a colonization defect. In E. coli deletion of xth and nfo leads to a more profound defect in DNA repair than either single mutant. Consistent with this observation, we found that an xth::Tn Δnfo double mutant showed a ∼10-fold defect in colonization that appears slightly greater than the ∼5-fold defect in colonization of the Δnfo mutant alone (Table 1), although this difference is not statistically significant for this number of replicates tested (p>0.05). Thus, these results suggest that BER is important for V. cholerae to colonize the infant mouse intestine when in competition. These results also show a critical function for Nfo in survival, which has not been apparent under laboratory conditions.
Loss of mismatch repair function has been shown to be either beneficial or detrimental depending on the pathogen studied [12], [13], [14], [15]. We found that a transposon mutant in mutS, which encodes the gene product that initially binds to a mismatch, resulted in a decrease in colonization efficiency (Table 1). We constructed a clean deletion of mutS (ΔmutS) to ensure the defect was not due to the transposon. We found that the clean deletion of mutS was also attenuated in its ability to colonize the intestine suggesting that mismatch repair or at least MutS is important for V. cholerae pathogenesis (Table 1). We also found that a second clean deletion of mutS showed a similar competitive index defect (CI = 0.18±0.03) suggesting that the colonization defect was not due to mutations in the first mutS clean deletion strain. We noted that the mutS transposon mutant was more defective than its clean deletion counterpart (Table 1). This difference may be due to a polar effect of the transposon or mutations acquired by the mutS::Tn strain during outgrowth of the original isolate. We also tested the colonization proficiency of a Δnfo ΔmutS double mutant and found that the colonization defect of this double mutant appears slightly greater than either the Δnfo or ΔmutS mutant alone (Table 1) although this difference is not statistically significant for this number of replicates tested (p>0.05).
The requirement of BER and MMR for V. cholerae to efficiently colonize the infant mouse intestine suggests that V. cholerae experiences DNA damage in the mouse, and that a reduced ability to repair such damage is detrimental for V. cholerae pathogenesis.
V. cholerae base excision repair and mismatch repair mutants show classic DNA repair defects
V. cholerae genes encoding Xth, Nfo and MutS were identified based on sequence similarity with their well-studied E. coli homologs. To ensure the V. cholerae homologs possessed their predicted functions we tested our mutant strains for the well characterized phenotypes described in other bacterial systems. Loss of mismatch repair causes an increase in mutation rate often referred to as a mutator phenotype [42]. We found that our ΔmutS mutant had a significantly increased mutation frequency compared with the wild type control (Figure 1A). The wild type phenotype could be restored by expression of mutS from a plasmid but not by the plasmid itself (Figure S1B). This result indicates that MutS in V. cholerae shares the same activity as its other well studied bacterial homologs in the repair of DNA replication errors.
Fig. 1. Phenotypes of V. cholerae DNA repair mutants. 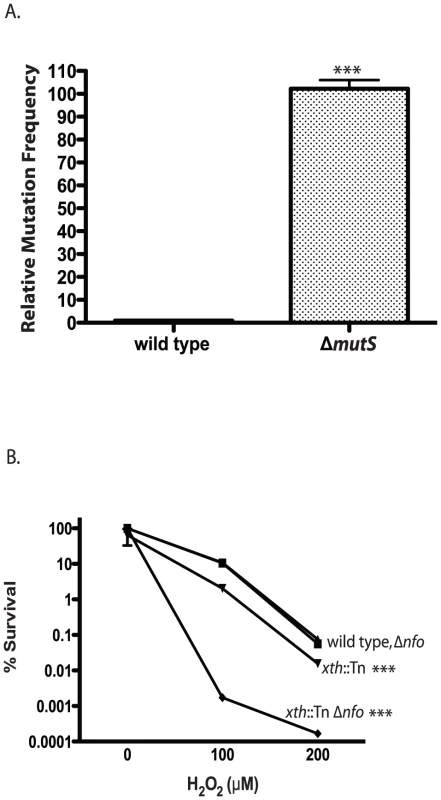
A. Mutation frequency. The number of ΔmutS rifampicin resistant colonies relative to wild type colonies is shown. The number of V. cholerae colonies was normalized to 1. The error bars reflect the SEM from at least 3 independent experiments (*** p<0.001). B. Hydrogen peroxide sensitivity. The sensitivity of wild type (▪), xth::Tn (▾), Δnfo (▴) and xth::Tn Δnfo (♦) strains to increasing concentrations of hydrogen peroxide are shown ± SEM from at least 3 independent experiments. The xth::Tn mutant is statistically different from the wild type and the Δnfo mutant at 100µM and 200µM H2O2 (*** p<0.001). The xth::Tn Δnfo strain is significantly different from the wild type, xth::Tn and Δnfo mutant at 100µM and 200µM H2O2 (*** p<0.001). Loss of Xth activity in E. coli renders the strain sensitive to hydrogen peroxide (H2O2) [43]. We found that our xth::Tn strain was also sensitive to H2O2 (Figure 1B). Loss of nfo activity alone does not greatly sensitize E. coli to H2O2 but loss of xth and nfo creates a strain with increased sensitivity to H2O2 [43]. We found a similar effect in V. cholerae where the xth::Tn Δnfo strain was much more sensitive to H2O2 then the xth::Tn mutant alone (Figure 1B). Furthermore, high level expression of nfo from a plasmid complemented the H2O2 sensitivity of the xth::Tn Δnfo mutant (Figure S1C). These results suggest that V. cholerae Nfo acts like its E. coli homolog.
Passage through the mouse increases the mutation frequency of V. cholerae
The requirement of BER and mismatch repair (MMR) systems for V. cholerae to efficiently colonize the mouse intestine suggests that V. cholerae experiences increased DNA damage while in the mouse. To address this possibility we measured the mutation frequency of V. cholerae following passage though the mouse as compared to passage in liquid culture. We inoculated five mice and five liquid cultures with the same size inoculums of V. cholerae. The following day we purified bacteria from the mouse intestine (see Materials and Methods). We plated both V. cholerae passaged through the mouse and grown in liquid cultures followed by selection for resistance to two antibiotics we used as an indicator for measuring mutation frequency. The first was a gain of function mutation in rpoB conferring resistance to rifampicin; the second was a loss of function of thyA conferring resistance to trimethoprim. Mutations in rpoB and thyA are well characterized markers for increases in mutation frequency [44], [45], [46]. We found that following passage of V. cholerae through the mouse there was an ∼2 fold increase in rifampicin resistance and ∼2.5 fold increase in trimethoprim resistance compared to the liquid culture grown strains (Figure 2A, B). We sequenced 19 trimethoprim resistance isolates that were passed through the mouse and 20 isolates obtained following growth in liquid culture. We identified 39 unique mutations in thyA (data not shown) suggesting that our results were not influenced by a mutation acquired early on in the procedure. We did not observe a bias in the types of mutation from the two conditions. These results suggest that passage through the mouse results in an increase in mutation rate for V. cholerae suggestive of an increase in DNA damage and the need for repair mechanisms.
Fig. 2. Mutation frequency of culture vs. mouse passaged wild type V. cholerae. 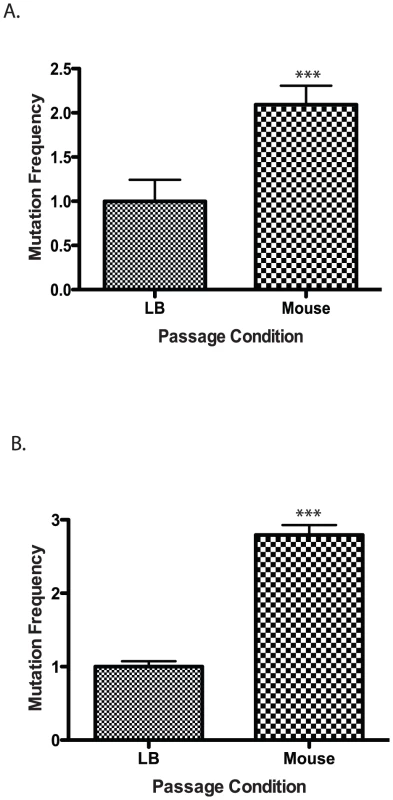
Wild type cells were grown in LB or passaged through a mouse and plated on (A) rifampicin or (B) trimethopirin to determine the number of resistant colonies. The results show the average mutation frequency of V. cholerae from 5 mice relative to the average mutation frequency from 5 LB grown cultures. The average mutation frequency of the LB grown V. cholerae cultures was normalized to 1. The error bars reflect the SEM from at least 3 independent experiments (*** p<0.001). Defense against RNS is required for V. cholerae to colonize the infant mouse
We have identified two DNA repair mechanisms required by V. cholerae to efficiently colonize the infant mouse, and have shown that V. cholerae passaged through a mouse has an increased mutation frequency. Thus, we sought to identify potential causes of DNA damage for V. cholerae while in the mouse to understand the requirement for BER and MMR in the mouse. A major source of DNA damage for intracellular pathogens is from host produced ROS and RNS. While V. cholerae is a non-invasive pathogen we considered that it still may experience ROS and RNS at some point during infection. We used a genetic approach to determine if ROS/RNS affected V. cholerae colonization and if so what type(s) of ROS/RNS were most important during this encounter.
Bacteria have several enzymes to detoxify ROS/RNS. Each enzyme detoxifies a specific type of ROS or RNS [for review see [18], [19]. For example catalases/peroxidases decompose H2O2, superoxide dismutases remove superoxide and ferrisiderophore reductases remove nitric oxide. Bacteria can contain multiple proteins capable of dealing with one type of stress. V. cholerae possesses two catalases/peroxidases (KatB/PerA) and one alkyl hydroperoxide reductase (AhpC), three superoxide dismutases (SodA/B/C) but only one ferrisiderophore reductase (HmpA). We tested mutants defective for each of these different types of defense enzymes to identify the type(s) of radicals that may be damaging V. cholerae in the mouse (Table 2).
Tab. 2. Ability of V. cholerae mutants defective in ROS or RNS detoxification to colonize the infant mouse intestine in competition with the parental strain (WT). 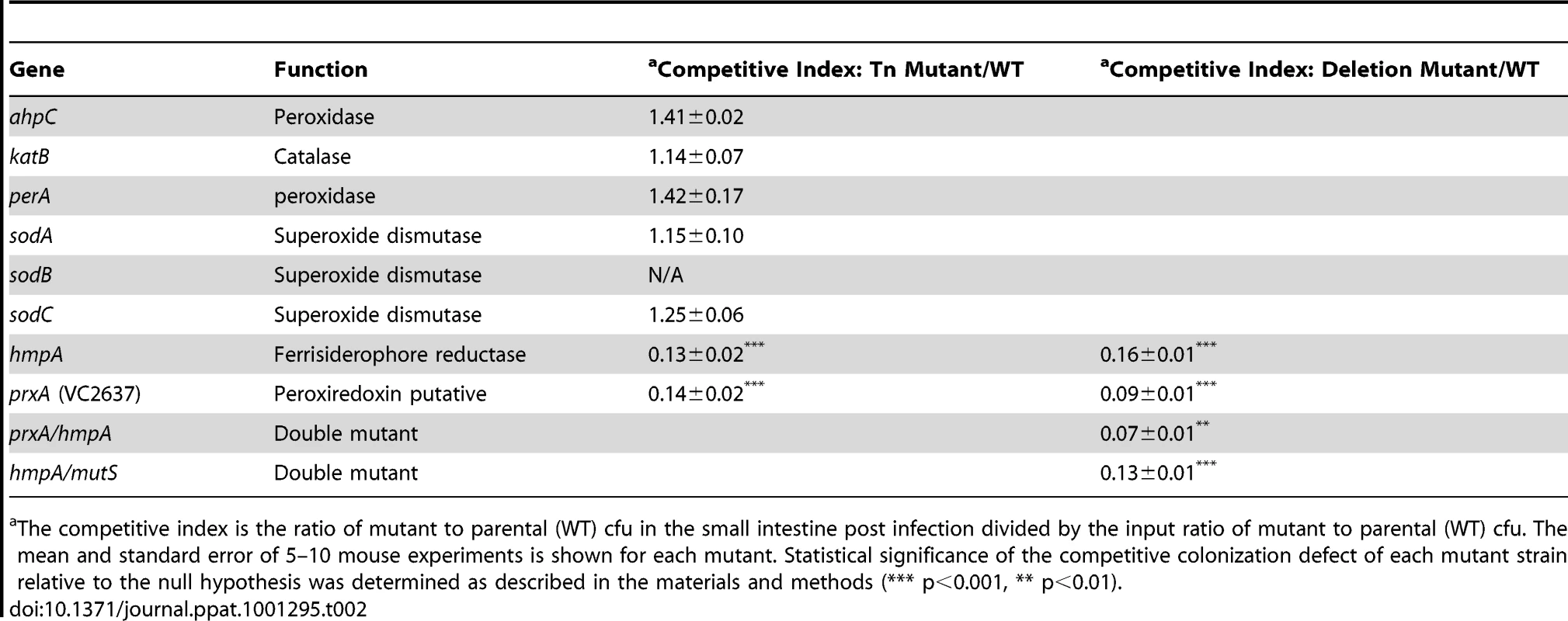
The competitive index is the ratio of mutant to parental (WT) cfu in the small intestine post infection divided by the input ratio of mutant to parental (WT) cfu. The mean and standard error of 5–10 mouse experiments is shown for each mutant. Statistical significance of the competitive colonization defect of each mutant strain relative to the null hypothesis was determined as described in the materials and methods (*** p<0.001, ** p<0.01). RNS, including nitric oxide, have been shown to be powerful antimicrobial agents. The most well studied RNS defense enzyme in bacteria is Hmp, a ferrisiderophore reductase that destroys nitric oxide [47]. V. cholerae carries an hmp homolog, hmpA. Both an hmpA::Tn mutant and a ΔhmpA deletion mutant showed a defect in colonizing the infant mouse intestine (Table 2). Deletion of hmpA delayed V. cholerae growth in the presence of a nitric oxide donor but not in the absence (Figure 3A) consistent with previous observations in other bacteria [20], [48]. This suggests that V.cholerae may encounter deleterious RNS during passage in the mouse. The growth defect of the ΔhmpA mutant in the presence of a nitric oxide donor could be complemented by ectopic expression of hmp from the arabinose inducible plasmid pBAD18 (Figure S1D). In fact, expression of hmp from pBAD18 allowed the ΔhmpA mutant to recover growth more rapidly than the parental strain in the presence of a nitric oxide donor.
Fig. 3. Growth of RNS/ROS mutants under stress. 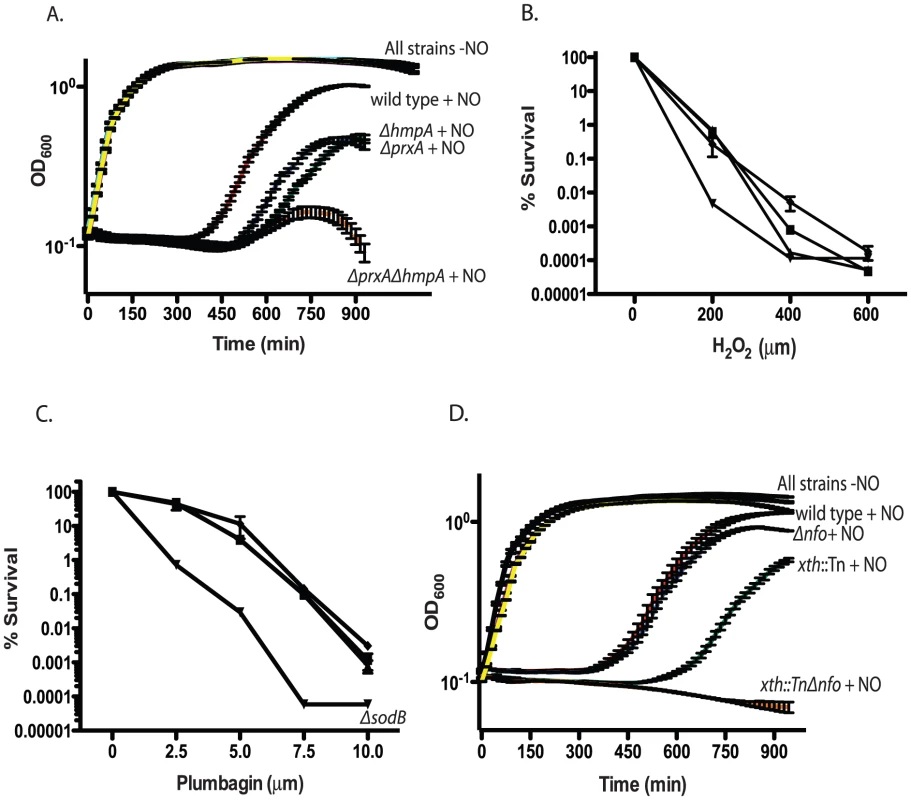
A. Nitric oxide stress. Exponentially growing cultures of wild type V. cholerae, the ΔhmpA mutant, ΔprxA mutant or ΔprxA ΔhmpA double mutant were grown with or without 1 mM spermine NONOate as a nitric oxide donor. The recovery and growth of each strain was monitored over time. The averages of 3 experiments are shown for each strain. Curves for wild type, ΔhmpA, ΔprxA and ΔprxA ΔhmpA double mutant treated with spermine NONOate are labeled as “+NO” for clarity. All strains grown in LB without spermine NONOate cluster together and are shown on the plot with the label “All strains−NO”. wild type+NO (red), ΔhmpA+NO (green), ΔprxA+NO (blue), ΔprxA ΔhmpA+NO (orange), wild type−NO (black), ΔhmpA−NO (purple), ΔprxA−NO (cyan) and ΔprxA ΔhmpA−NO (yellow). The growth of the ΔprxA, ΔhmpA and ΔhmpA ΔprxA mutant is significantly delayed by NO compared to wild type by 435 min (p<0.05). B. H2O2 sensitivity. Wild type (▪) and katB::Tn (▴), perA::Tn (▾) and ahpC::Tn (♦) mutants were plated on increasing concentrations of hydrogen peroxide. Cfu were determined after overnight growth. The average of 3 experiments is shown. C. Superoxide sensitivity. Wild type (▪) and sodA::Tn (▴), ΔsodB (▾) and sodC::Tn (♦) mutants were plated on agar containing increasing concentration of the superoxide generating compound plumbagin. Cfu were determined after overnight growth. The average of 3 experiments is shown. D. Exponentially growing cultures of wild type, the xth::Tn mutant, Δnfo mutant or xth::Tn Δnfo double mutant were grown with or without 1 mM spermine NONOate as a nitric oxide donor. The recovery and growth of each strain was monitored over time. The averages of 3 experiments are shown for each strain. Curves for wild type, xth::Tn, Δnfo and xth::Tn Δnfo treated with spermine NONOate are labeled as “+NO” for clarity. All strains grown in LB without spermine NONOate cluster together and are shown on the plot with the label “All strains−NO”. wild type+NO (red), xth::Tn+NO (green), Δnfo+NO (blue), xth::Tn Δnfo+NO (orange), wild type−NO (black), xth::Tn−NO (purple), Δnfo−NO (cyan) and xth::Tn Δnfo−NO (yellow). At 750 min the growth delay of the xth::Tn mutant compared to the wild type in NO is significant (p<0.001) while the growth delay of the Δnfo mutant is not. After testing all previously predicted antioxidant enzymes we began to mine the V. cholerae genome for additional putative antioxidant enzymes. We began by searching for putative proteins that belonged to large antioxidant families. Enzymes, such as AhpC, belong to the Peroxiredoxin (PRX) family. Searching for peroxiredoxin family proteins yielded a putative defense enzyme we have called PrxA (VC2637). PrxA, classified is a peroxiredoxin-5 family protein, is found in several pathogenic bacteria and is a distant homolog of a macrophage peroxynitrite detoxification protein [49].
Deletion of prxA did not effect V. cholerae growth in LB alone but significantly delayed V. cholerae growth in the presence of a nitric oxide donor (Figure 3A). Furthermore, both the prxA::Tn mutant and the ΔprxA allele we constructed caused a decrease in the ability of V. cholerae to colonize the infant mouse in competition assays (Table 2). The growth defect of the ΔprxA mutant in the presence of a nitric oxide donor could be complemented by ectopic expression of prxA from the arabinose inducible plasmid pBAD18 (Figure S1E). The discovery of a new gene required for both defense against RNS and efficient colonization of the infant mouse further supports our findings that V. cholerae may be exposed to RNS during passage though the mouse.
We tested the sensitivity of a ΔprxA ΔhmpA double mutant and found that the growth of the double mutant in the presence of a nitric oxide donor was even more delayed than either the ΔprxA or ΔhmpA single mutant alone (Figure 3A). We also tested the colonization efficiency of a ΔprxA ΔhmpA (Table 2) and found that it was not significantly less than the ΔprxA mutant alone (p>0.05). Thus hmpA and prxA are both important for colonization but the effects were not additive.
We asked if defects in ROS defense also affect V. cholerae colonization. Disruption of ahpC, katB, perA, sodA or sodC did not affect the ability of V. cholerae to colonize the infant mouse and these deficiencies did not affect the ability of V. cholerae to colonize the infant mouse in competition experiments (Table 2). We did not test the sodB::Tn mutant since both it and a ΔsodB deletion strain we constructed had a decreased plating efficiency and grew very poorly compared to the parental strain (Figure S1F). Thus, while SodB appears to be important for growth of V. cholerae under laboratory conditions we did not pursue the sodB mutant in mouse experiments.
Interestingly, of ahpC, katB and perA only disruption of perA sensitized V. cholerae to H2O2 in vitro (Figure 3B). Furthermore, of strains disrupted individually for sodA, sodB and sodC only disruption of sodB sensitized V. cholerae to the superoxide generating compound plumbagin (Figure 3C). We also tested the ΔprxA mutant but found that it did not show increased sensitivity to either H2O2 or plumbagin (Figure S2A and data not shown). It is possible that some of these known ROS defense enzymes overlap in function masking the effects of a deficiency in any one gene in vitro or in mouse studies. For other bacterial pathogens and symbionts deletion of several or all catalases and superoxide dismutases has been required before a strong effect on virulence or symbiosis was observed [50], [51], [52]. Currently, the results from our analysis suggest RNS may pose a significant barrier to V. cholerae in colonizing the infant mouse. ROS may also play a role, however their effect is not immediately evident in our analysis.
XthA appears to be more important than Nfo in protecting V. cholerae against environmental stress in vitro (Figure 1B), yet Nfo appears to be more important for colonization of the small intestine (Table 1). The requirement for ΔhmpA and ΔprxA for efficient intestinal colonization lead us to ask if Nfo was required for defense against nitric oxide. We monitored the growth of the xth::Tn, Δnfo and xth::Tn Δnfo mutants in the presence of a nitric oxide donor. We found that, at least in vitro, xth:Tn was more important than Nfo for protection against nitric oxide (Figure 3D). We also found that the double mutant was again more sensitive to the stress than either single mutant alone (Figure 3D).
Colonization defective mutants have increased sensitivity to acidified nitrite
Our results led us to ask if our DNA repair and RNS defense defective V. cholerae mutants were sensitive to any host defenses. The intestine has several innate defenses [37]. We tested many of these defenses including lysozyme, phospholipase, antimicrobial peptides, complement, bile, changes in osmolarity and pH, however, we did not observe any difference in sensitivity between the parental and the mutant strains (data not shown). RNS have been shown to be generated by macrophages to kill phagocytised bacteria [reviewed in [53]]. The RNS from macrophages is generated by an inducible nitric oxide synthase (iNOS). Inhibition of iNOS activity has been shown to rescue the virulence defects in hmp mutant strains of Salmonella enterica serovar typhimurium [20]. However, our hmpA::Tn V. cholerae mutant showed no difference in it ability to colonize the intestine of a wild type or isogenic iNOS−/−infant mouse (Table S1).
Thus, our results suggest that the colonization defect of the DNA repair and RNS defense mutants may occur before V. cholerae is exposed to the host defenses found in the small intestine. In the stomach V. cholerae is exposed to low pH in combination with µM amounts of nitrite from ingested food and the salivary nitrite cycle [25], [26], [27]. Acidified nitrite produces a variety of toxic RNS. We quantified the amount of nitrite in the infant mouse stomach using the Griess reaction and found that it was 20.0±0.7 µM, which is similar to that of humans [26]. The pH range of human gastric juice is reported as 1–3 [23], [24]. We determined the pH of the infant mouse stomach to be 4.5±0.1 using a fluorescent pH sensitive dye. This measurement is conservative and the pH of the infant mouse gastric juice may be even less (see Materials and Methods). Thus, the infant mouse stomach is sufficiently acidic to promote the formation of acidified nitrite.
At pH 3 in rich medium we found that V. cholerae had a greater than 99.9% decrease in survival in less than 1 minute (data not shown) agreeing with similar work examining V. cholerae acid tolerance [54]. We did not find a difference in survival between the parental and mutant strains at low pH (1–4) levels (data not shown). We gradually increased pH to identify the lowest level at which V. cholerae could grow. At pH 5.5 V. cholerae and the DNA repair and RNS defense mutants grew with identical kinetics (Figure 4A). We titrated nitrite into the growth medium and found that nearly all the mutant strains showed a growth defect compared to the wild type at pH 5.5 in the presence of 400 µM nitrite (Figure 4B). No differences in growth between wild type and mutant strains were observed at pH 7.0 with or without 400 µM nitrite (Figure S3A, B). Not only did low pH and nitrite slow the growth of our mutants but the ΔhmpA, ΔprxA, Δnfo and xth::Tn Δnfo mutants began to show a decrease in optical density after longer exposure (Figure 4B) suggesting the cells were lysing.
Fig. 4. Effect of nitrite on growth of wild type and mutant V. cholerae strains. 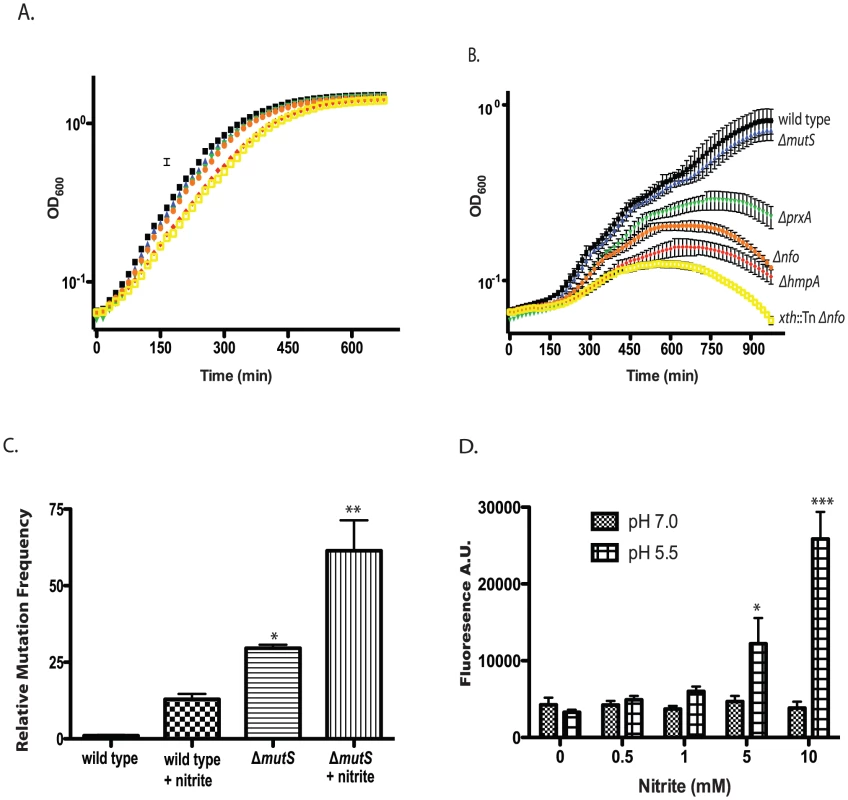
A/B. Exponentially growing cultures of wild type and ΔmutS, Δnfo, ΔprxA, ΔhmpA and Δnfo xth::Tn mutants were grown in LB buffered at pH 5.5 in the absence (A) or presence (B) of 400 µM sodium nitirite. The average of three experiments is shown for each strain. Wild type (black squares), ΔmutS (blue triangles), Δnfo (orange circles), ΔprxA (green inverted triangle), ΔhmpA (red diamond) and Δnfo xth::Tn (yellow open square). C. Mutation frequency as measured by rifampicin resistant colony formation frequency from wild type and ΔmutS mutant cultures grown at pH 5.5 in the presence or absence of 600 µM sodium nitrite. The average mutation frequency of the wild type grown in the absence sodium nitrite was normalized to 1 (* p<0.05, ** p<0.01 compared to wild type) D. Intracellular RNS production following nitrite treatment. Wild type cultures were grown at pH 7.0 or 5.5 plus 0, 0.5, 1.0, 5.0, or 10.0 mM sodium nitrite. After washing cells were exposed to the radical binding dye H2DCFDA. After removal of media, cells were lysed and H2DCFDA fluorescence was measured. The average of at least 3 independent experiments is shown with error bars representing the SEM (* p<0.05, ** p<0.01). Only the growth of the ΔmutS strain was unaffected at 400 µM. We considered that while MutS may not be required for survival of acidified nitrite during this time course it may be required to prevent acidified nitrite induced mutations in V. cholerae that are detrimental for colonization. We grew V. cholerae in LB at pH 5.5 over night in the presence or absence of 600 µM nitrite and then plated for rifampicin resistant colonies. We found that V. cholerae grown in the presence of nitrite had a greater than 10-fold increase in mutation frequency compared to the media only control (Figure 4C). Loss of MutS then increased the mutation frequency of V. cholerae in nitrite at pH 5.5 ∼ an additional 5-fold (Figure 4C). Thus, MutS may be important to prevent acidified nitrite induced mutations that could impair the ability of V. cholerae to colonize the infant mouse. To further test this possibility we created a ΔhmpA ΔmutS double mutant and tested its colonization proficiency (Table 2). Interestingly, the colonization defect of the ΔhmpA ΔmutS double mutant was not significantly different than either of the single mutants alone (p>0.05). This result may suggest that HmpA and MutS may share a similar defense pathway in the infant mouse.
Additionally, E. coli MutS can recognize an O6-methyl-dG:dC base pair, a mutation which can occur by alkylation of G bases [55]. Therefore it is possible that MutS may also be important for protection again some type of alkylation that occurs in the mouse stomach.
Low pH and nitrite induce radical formation in V. cholerae
If acidified nitrite produces RNS that damaged V. cholerae DNA, we reasoned that we should be able to detect increased intracellular radical formation in V. cholerae following nitrite treatment. We grew V. cholerae at pH 5.5 with increasing amounts of nitrite and assayed for radical formation using 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA). H2DCFDA is a cell permeable dye that fluoresces after reacting with RNS and ROS species. H2DCFDA reacts with several ROS and RNS including hydrogen peroxide, nitric oxide and peroxynitrite [56], [57], [58]. We found that H2DCFDA fluorescence correlated with increasing nitrite concentration at pH 5.5 indicating an increase in intracellular radicals (Figure 4D). For comparison, we found that H2DCFDA fluorescence did not increase over the concentrations of nitrate examined at pH 7.0. These results further support the requirement for low pH to induce radical formation from nitrite sources (Figure 4D).
A colonization defect is observed for the DNA repair and RNS defense mutants early after inoculation
Our results suggested that V. cholerae may experience DNA damage as it passes through the stomach. If so, we were curious if a colonization defect could be observed at an early time point after inoculation. We repeated the competitive colonization assays testing the ΔmutS, Δnfo, ΔprxA and ΔhmpA mutants. We found that at 3 hours post inoculation each mutant strain already showed a 50–60% colonization defect when co-inoculated with the wild type (Table S2) though this defect was not as great as the 5–20% defects reported in Table 1. This suggests that a defect of the mutant strains is detrimental for colonization early after inoculation. Angelichio et al. [59] reported that V. cholerae populations in the small intestine do not show a significant increase in number until between 10–24 h post inoculations. These results suggest that the effects of the DNA damage are ∼50% detrimental at the earliest stages of infection and become more apparent as the bacteria replicate to high numbers in the intestine. Such a conclusion is consistent with the concept of damage occurring primarily in the stomach.
The colonization defect of the DNA repair and RNS defense mutants is rescued by neutralizing stomach acid
Our results support a relationship between acidified nitrite sensitivity and colonization defects in our RNS and DNA repair. We reasoned that if acidified nitrite in the stomach was responsible for the colonization defects of our mutants then neutralizing stomach acid in the mouse should relieve, at least in part, the observed colonization defects. We used sodium bicarbonate to neutralize the mouse stomach acid. When we inoculated infant mice with our DNA repair and RNS defense defective mutants in the presence of sodium bicarbonate all four mutant strains (Δnfo, ΔmutS, ΔprxA and ΔhmpA) showed significant improvement in their ability to colonize the intestine in competition with the parental strain (Table 3). In fact the colonization defect of the ΔmutS and Δnfo mutant was completely rescued. The colonization defect of the ΔprxA was restored to near wild type levels. The colonization defect of the ΔhmpA mutant was partially rescued although this difference is not statistically significant for the number of replicates tested (p>0.05).
Tab. 3. Effect of NaHCO3 on mutant V. cholerae ability to colonize the infant mouse intestine in competition with the parental strain (wild type). 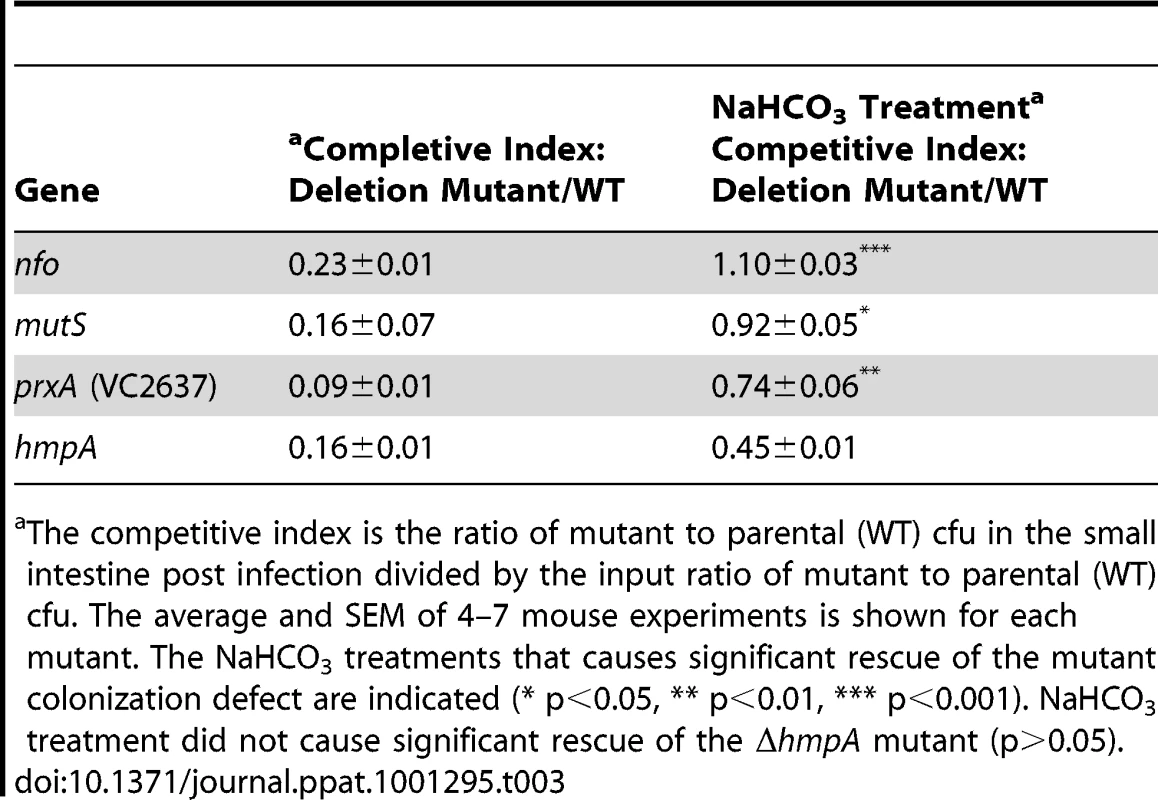
The competitive index is the ratio of mutant to parental (WT) cfu in the small intestine post infection divided by the input ratio of mutant to parental (WT) cfu. The average and SEM of 4–7 mouse experiments is shown for each mutant. The NaHCO3 treatments that causes significant rescue of the mutant colonization defect are indicated (* p<0.05, ** p<0.01, *** p<0.001). NaHCO3 treatment did not cause significant rescue of the ΔhmpA mutant (p>0.05). The nitrite concentration of the infant mouse stomach after sodium bicarbonate treatment remained nearly unchanged at 20.3±0.8 µM.
Discussion
We have shown that V. cholerae must defend against DNA damage to efficiently colonize the infant mouse intestine and that such damage likely occurs early during infection as V. cholerae enters the stomach. We have demonstrated that V. cholerae specifically requires BER and MMR pathways to efficiently colonize the infant mouse intestine. Furthermore, we have identified one previously known and one novel RNS defense protein that facilitates intestinal colonization of the infant mouse. These DNA repair and RNS defense proteins were also required for V. cholerae to grow or maintain genomic fidelity in the presence of acidified nitrite. Furthermore the colonization defects of each mutant could be partially or fully complemented by neutralizing stomach acid suggesting that RNS defense and DNA repair share a common defensive role in the mouse.
V. cholerae has been shown to be very sensitive to low pH [54]. For this reason, human volunteers have their stomach contents neutralized to promote experimental V. cholerae infection as is done with live attenuated vaccine studies [60]. In the recently developed infant rabbit model for cholera, stomach acid is also neutralized and cimetidine is administered to prevent re-acidification in order for V. cholerae to colonize the infant rabbit intestine [61]. However, our DNA repair and RNS defense mutants did not show increased sensitivity to low pH compared to the parent strain. Agreeing with our observations a large screen used to identify genes necessary for colonization and tolerating low pH in V. cholerae did not identify any of the genes we reported here for influencing colonization [62]. While low pH of the stomach alone is undoubtedly detrimental towards V. cholerae our results suggest that neutralizing stomach acid may also be important to prevent RNS formation by acidified nitrite. We propose that the defect in colonization of the DNA repair and RNS defense mutants is due to RNS formation.
Acidified nitrite is present in the stomach where gastric juice interacts from nitrite sources from the diet or salivary nitrite pathway. The chemistry of acidified nitrite is known to produce several potentially deadly radicals (1).(1)Both NO• and NO2• can directly attack cellular macromolecules, but they can also interact with other radicals to form further species such as peroxynitrite and hydroxyl acids. From equation (1) we can see that a key factor in this process is the pH of the solution. In the human stomach, normal nitrite concentrations range from 10–50 µM [24] but at a pH of 1–3 acidification and radical production can happen rapidly. The low pKa of HNO2 may explain in part why we required much higher concentrations of nitrite to observe detrimental effects on the mutants we studied. pH 5.5 was the lowest pH level we could successfully grow V. cholerae, a value well above the pKa of HNO2. Thus at pH 5.5 the acidification of nitrite would occur more slowly and higher concentrations of nitrite would be necessary for mass action to drive the acidification and radicalization of nitrite. Agreeing with this we did not observe any effect of nitrite on the growth of our mutants or V. cholerae at pH 7.0. While acidified nitrite has been shown to effectively kill several bacterial pathogens [28], [29], [30], [31], the mechanism of its action and bacterial defenses to protect against it have remained unknown. We have now shown that MutS, Nfo, HmpA and PrxA are required for protection of acidified nitrite in V. cholerae.
While our results support a role for acidified nitrite in the stomach acting as a major DNA damaging agent, we have not identified the exact location in the gastrointestinal track where the damage occurs. The most apparent location is the stomach where ingested V. cholerae mixes with gastric juice. However it is possible that DNA damage induced by acidified nitrite radicals occurs, or continues to occur, in the upper intestinal tract. As gastric juice exits the stomach it is neutralized by bile salt, etc. and can reach a pH close to 8 [24]. Since the stability of at least some RNS, such as peroxynitrite, increases with increasing pH [63] the upper intestinal tract may provide a more favorable environment for RNS to reach V. cholerae and induce DNA damage.
Bicarbonate has been shown to induce V. cholerae virulence genes in a ToxT dependent fashion [64]. Abuaita and Withey show that significant upregulation of both cholera toxin and tcpA gene expression are observed 3–4 h after addition of bicarbonate [64]. While our inoculation of V. cholerae occurs on a much shorter time scale (∼5–15 min after exposure of bicarbonate) and the majority of V. cholerae has passed into the small intestine before 3 h, it is possible that some bicarbonate induced gene regulation may also aid in the bicarbonate rescue of the colonization defect of our DNA repair mutants.
While the debate over the benefits and detriments of increased mutation frequency for pathogenesis continues, we have shown that increased mutation frequency is detrimental to V. cholerae pathogenesis, at least for the short-term colonization of the infant mouse intestine. However, we cannot exclude the possibility that increased mutation frequency affects long-term survival of V. cholerae in the host. After the initial decrease in competitiveness it is possible that increased mutation frequency in V. cholerae could make it more competitive in later stages of colonization or during release into the environment. It would be interesting to test multiple clinical isolates for a mutator phenotype to address this question.
The Xth and Nfo homologs of V. cholerae have strong sequence similarity to their E. coli counterparts. We have shown that V. cholerae and E. coli deletion mutants of xth and nfo also share a similar pattern of sensitivity to hydrogen peroxide. Nfo and Xth have been most extensively studied in E. coli. In E. coli Xth is responsible for greater than ∼90% of all AP endonuclease activity in the cell [39], [40]. In E. coli, an xth mutant is very sensitive to a variety of DNA damaging agents whereas nfo mutants generally show milder effects [41]. Interestingly, we have shown that in V. cholerae Nfo is more important for colonization of the infant mouse than Xth. Our nfo xth double mutant suggests that Xth may play a role in colonization when Nfo is absent. However, for whatever damage is occurring, Nfo appears to play a more important role in the mouse.
It is possible that Nfo and Xth are also used differentially for repair of specific types of lesions. Preferences for specific types of damaged bases between Nfo and Xth from E. coli have been previously reported [65]. If RNS are responsible for DNA damage in the mouse we suggest that Nfo may have enhanced ability to aid in the repair of nitrosylative base damage. Additionally, there may be differential expression of xth and nfo or their preferential glycosylase partners in the host.
In our efforts to identify ROS and RNS defense enzymes required for intestinal colonization, we identified a new protein we have called PrxA that was required for RNS defense. Until now Hmp has been the only bacterial protein identified to detoxify RNS, specifically nitric oxide. We have shown that like HmpA, PrxA protects V. cholerae against the nitric oxide donor spermine NONOate. Similarly, HmpA and PrxA both protect V. cholerae against acidified nitrite. This agrees with previous work showing that Hmp protects Salmonella against nitric oxide and acidified nitrite [34]. While the species(s) produced by acidified nitrite that HmpA and PrxA defend against is not clear, we presume that it is a RNS. PrxA homologs are not as prevalent in bacteria as HmpA homologs, but they are found in several pathogens including Yersina pestis, Haemphilus influenza and Neisseria gonorrhoeae. It will be interesting to determine if PrxA homologs share a similar RNS defense role in other bacteria.
Our observation that HmpA and PrxA are required for colonization lead us to suggest that V. cholerae encounters RNS stress during infection. When studying ROS defense genes we found that deletion of SodB was detrimental for normal V. cholerae growth. This indicated that normal growth of V. cholerae must generate a significant amount of superoxide managed by SodB. The growth defect of the ΔsodB mutant prevented us from analyzing it by competition in the mouse model. While we did not identify any other single ROS defense enzyme that affected intestinal colonization it is possible that construction of various double mutants may show that V. cholerae must also deal with ROS during disease progression. In bacterial pathogens where SODs have been shown to be necessary for virulence, it is generally the periplasmic SOD that is required as this SOD encounters superoxide entering the cells from the environment [16], [66]. However, the V. cholerae periplasmic SOD, SodC, was not required for intestinal colonization suggesting V. cholerae does not experience superoxide stress from the host.
Materials and Methods
Ethics statement
The animal experiments were performed with protocols approved by Harvard Medical School Office for Research Protection Standing Committee on Animals. The Harvard Medical School animal management program is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care, International (AAALAC), and meets National Institutes of Health standards as set forth in the Guide for the Care and Use of Laboratory Animals (DHHS Publication No. (NIH) 85-23 Revised 1996). The institution also accepts as mandatory the PHS Policy on Humane Care and Use of Laboratory Animals by Awardee Institutions and NIH Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training. There is on file with the Office of Laboratory Animal Welfare (OLAW) an approved Assurance of Compliance (A3431-01).
Bacterial strains
Strains and plasmids are listed in Supporting Table S3. V. cholerae El Tor biotype strain C6706 and a spontaneous lacZ − derivative of C6706, were used as parental (wild type - WT) strains. E. coli DH5α λpir and Sm10 λpir were used for cloning and conjugation, respectively. Antibiotic concentrations used were streptomycin (Sm: 100 µg/ml or 500 µg/ml), kanamycin (Kan: 50 µg/ml), carbenicillin (Carb: 75 µg/ml) and chloramphenicol (Cm: 2.5 µg/ml for C6706 and 10 µg/ml for E. coli DH5α λpir). LB was used for all growth conditions [10 g/liter of tryptone (Bacto), 5 g/liter of yeast extract (Bacto), and 5 g/liter of NaCl] and was supplemented with 16 g/liter of agar (Bacto) for growth on plates. Arabinose was used at 0.1% for complementation assays. All ID numbers/ Accession numbers/for genes highlighted in this study are shown in Table S5.
DNA manipulations
The genomic sequence of C6706 has not been completed. We used the sequence of the close relative, N16961, for clone construction. For in-frame gene deletions of nfo, mutS, hmpA and prxA, genomic DNA surrounding the respective gene was amplified by crossover PCR and cloned into pWM91 for subsequent sacB-mediated allelic exchange in V. cholerae, as described [67], [68]. For complementation constructs, the respective gene was amplified from chromosomal DNA and cloned into plasmid pBAD18 after digestion with KpnI and SalI. The respective gene was induced by adding arabinose to the growth medium. All cloning products were sequence-verified, and the nucleotide sequence of all primers used is listed in Table S4.
Infant mouse colonization competition assays
A modified version of the protocol of Baselski and Parker [69] was performed for infection and recovery of C6706 derived strains. C6706 or C6706 lac− and mutant strains were grown on LB-agar plates with Sm overnight at 37°C. Wild type and mutant strains were mixed together in LB. 50 µl of this competition mixture (∼50 000 bacteria) was inoculated into a 5-day-old CD1 mouse pup (Charles River Company). Serial dilutions of the competition mixture were plated in LB+Sm100 and enumerated to determine the input ratio of wild type and mutant strain. After incubation at 30°C for 3 h or 18 h the mouse pups were sacrificed and small intestines were removed and homogenized in 10 ml of LB. Serial dilutions were plated in LB+Sm100 and enumerated to determine the output ratio of wild type and mutant strain. The competitive index for each mutant is defined as the input ratio of mutant/wild type strain divided by the output ratio of mutant/wild type strain. A minimum of four mice were assayed for each mutant strain. The in vivo experiments for the transposon and clean deletion strains were the accumulation of results performed on different days. For ease of communication we reported the average competitive index.
For NaHCO3 recue experiments, mice pups were first inoculated with 50 µl of 2.5 g/100 mL NaHCO3. After 3 h the pups were inoculated with 50 µl of the competition mixture in 2.5g/ 100 mL NaHCO3. iNOS−/− (#002609) and control C57BL/6J (#000664) mice were purchased from The Jackson Laboratory.
Mutation frequency assays
Rifampicin resistance assays
i) For ΔmutS mutation frequency and complementation assays cultures were grown to saturation at 37°C in LB Sm500 or Sm100. 500 µl of culture was plated on LB agar+50 µg/mL rifampicin. After overnight growth at 37°C rifampicin resistant colonies were scored. ii) For mouse passaged assays, 5 day old mouse pups were inoculated with 50, 000 cells of wild type V. cholerae. After incubation at 30°C for 18 h the mouse pups were sacrificed and their small intestines removed and homogenized in 10 ml of LB+Sm500. The 10 ml of homogenized intestine was passed through cheese cloth and a 3.1µm filter. This filtering retained >90% of V. cholerae and removed the majority of eukaryotic materials as determined by western blot against mouse actin (data not shown). We recovered ∼250 000–500 000 V. cholerae cfu per small intestine. The filtrate was grown to saturation. For the control experiment 50 000 wild type V. cholerae were inoculated into 10 mL of LB+Sm500 and grown to saturation. We then plated an equal number of cfu from both mouse passaged and control cultures on LB agar+50 µg/mL rifampicin and scored resistant colonies. Control mouse samples in which no V. cholerae had been inoculated did not grow in LB+Sm500. Primers used for sequencing rpoB are shown in Supporting Table S3.
Trimethoprim resistance assays
A modified version of the Belfort and Pedersen-Lane protocol [44] was used for identified trimethoprim resistant colonies. For mouse passaged trimethoprin assays, 5 day old mouse pups were inoculated with 50 000 cells of wild type V. cholerae. After incubation at 30°C for 18 h the mouse pups was sacrificed and their small intestines removed and homogenized in 10 ml of LB Sm500+50 µg/mL thymine. The 10 ml of homogenized intestine was passaged through cheese cloth and a 3.1µm filter. This filtering retained >90% of V. cholerae and removed the majority of eukaryotic materials as determined by western blot against mouse actin (data not shown). We recovered ∼250 000–500 000 wild type V. cholerae cfu per small intestine. The filtrate was grown to saturation. For the control experiment 50 000 wild type V. cholerae were inoculated into 10 mL of LB Sm500+50 µg/mL thymine and grown to saturation. We then plated an equal number of cfu from both mouse passaged and control cultures on M9 agar+0.1% CAS, 0.2% glucose, 50 µg/mL thymine and 20 µg/mL trimethoprim. After overnight growth at 37°C trimethoprim resistant colonies were scored. Control mouse samples in which no V. cholerae had been inoculated did not grow in LB+Sm500. The nucleotide sequences of the primers used for sequencing thyA are shown in Supporting Table S4.
To calculate the relative mutation frequency we plated equal numbers of cfu for both mouse and passaged and control samples. We calculated the average and standard error for the mutation rate for the control samples. Next we normalized the individual mutation frequencies from our 5 mice passaged samples and 5 control samples to the average control sample mutation frequency. This normalized the average control sample mutation frequency to 1 and showed the relative mutation frequency increase in mouse passaged samples.
Stomach pH and nitrite concentration determination
We have developed a fluorescence based assay to determine the pH of the infant mouse stomach. We first determined a standard curve using the fluorescent pH indicator Yellow/Blue DND-160 (Invitrogen) over a range from pH 3–8. We then extracted the gastric juice from 5 individual mice, diluted the sample 1∶2 with ddH2O (pH 7), added Yellow/Blue DND-160 and determined the fluorescence of the solution. Comparing these fluorescent values to our standard curve we determined the pH of the infant mouse stomach to be 4.5±0.1. We also note that this is a conservative measurement. In order to obtain enough liquid we diluted the gastric sample ∼1∶2 with ddH20 that was at ∼pH 7. Thus while water is not a buffer, the dilution of the gastric juice likely raised the final pH of our measurements.
Nitrite concentration was determined using the Griess Reagent System (Promega TB229). The concentration shown is the average of 10 mice treated with or without sodium bicarbonate.
Hydrogen peroxide sensitivity assays
Strains were grown to exponential phase in LB with Cm when required. Strains were serial diluted and spotted on LB plates containing increasing concentrations of hydrogen peroxide and incubated at 37°C overnight. For complementation Cm and arabinose were added while strains were growing in liquid, as well as in the LB agar plates.
Nitric oxide sensitivity growth curves
Strains were grown to exponential phase in LB. Strains were then diluted to OD600 0.01 in LB±1.0 mM spermine NONOate and grown at 37°C in a 96 well plate with aeration (SpectraMax Plus 384, Molecular Devices). OD600 readings were taken every 15 min.
Growth in acidified nitrite
Overnight cultures were diluted into LB and grown to log phase at 37°C with aeration. Cultures were diluted to OD600 0.05 in 25 mM MES buffered LB of pH 7.0 or 5.5 with or without the addition of 400 µM sodium nitrite (Sigma-Aldrich). The LB media and MES were adjusted to a pH of 7.0 and 5.5 (Corning pH meter 240) with additions of HCl, and filter sterilized (0.22 µm, Corning) prior to use. The growth of strains under various treatments were determined by OD600 measurement using a 96 well format spectrophotometer (SpectraMax Plus 384, Molecular Devices). Environmental parameters were set to 37°C with shaking and readings were taken every 15 minutes for 16 hours. Studies were conducted in quadruplicate.
Fluoroscein assay
Overnight cultures were diluted into 100 mL LB with Sm100 and grown to OD600∼0.8 (37°C, aeration). 10 mL of culture was dispensed into 15 mL conical and centrifugated at 5,000 RPM for 5 minutes. The supernatant discarded and cells resuspended in an equal volume of 25 mM MES buffered LB of pH 7.0 or 5.5 with or without the addition of sodium nitrite (500 µM, 1 mM, 5mM, or 10 mM). Cells were treated for 1.5 hours at 37°C with aeration then centrifugated at 5,000 RPM for 5 minutes at 4°C. The supernatant was discarded, cells resuspended in 1 mL PBS (LONZA), and transferred to a 1.5 mL eppendorf tube. The cells were centrifugated and washed an additional two times in 1× PBS before being resuspended in 1 ml of PBS with 10 µM 2′,7′-dichlorodihydrofluorescein diacetate (Molecular Probes, Invitrogen). The cells were incubated at room temperature for 30 minutes then centrifugated and washed three times to remove all free, extracellular dye. The cells were lysed in 225 µL of lysis buffer (MilliQ water with 0.1M EDTA) via sonication. Cell lysates were centrifugated at 15,000 RPM for 5 minutes, supernatant transferred to another 1.5 mL eppendorf tube and centrifugated again. Fluorescence was measured at 490 nm / 519 nm (excitation/emission) (SpectraMax Gemini XS). Fluorescence was normalized against protein concentrations, as determined by Bradford assay. Studies were conducted in triplicate.
Statistical methods
Statistical significance was assessed for mouse colonization assays and ΔmutS mutation frequency assays using a one-way analysis of variance (ANOVA) using a Bonferroni post test to determine significant differences in competitive index between all pairs of V. cholerae mutants used in our study. Statistical significance of acidified nitrite, nitric oxide and H2O2 sensitivities was assessed using a mixed model, repeated measures two-way analysis of variance (ANOVA), generating a p value for each pair wise curves over the concentration range of H2O2 to determine the significance of our results. Statistical significance of rifampicin and trimethoprim resistant mutants from LB vs. mouse passaged samples were assessed using a paired t-test. Differences were considered significant at p<0.05. All calculations were performed using Graphpad Prisim version 5.
Supporting Information
Zdroje
1. CarpenterEP
CorbettA
ThomsonH
AdachaJ
JensenK
2007 AP endonuclease paralogues with distinct activities in DNA repair and bacterial pathogenesis. Embo J 26 1363 1372
2. MerinoD
Reglier-PoupetH
BercheP
CharbitA
2002 A hypermutator phenotype attenuates the virulence of Listeria monocytogenes in a mouse model. Mol Microbiol 44 877 887
3. O'RourkeEJ
ChevalierC
PintoAV
ThibergeJM
IelpiL
2003 Pathogen DNA as target for host-generated oxidative stress: role for repair of bacterial DNA damage in Helicobacter pylori colonization. Proc Natl Acad Sci U S A 100 2789 2794
4. RichardsonAR
SolivenKC
CastorME
BarnesPD
LibbySJ
2009 The Base Excision Repair system of Salmonella enterica serovar typhimurium counteracts DNA damage by host nitric oxide. PLoS Pathog 5 e1000451
5. SuvarnapunyaAE
LagasseHA
SteinMA
2003 The role of DNA base excision repair in the pathogenesis of Salmonella enterica serovar Typhimurium. Mol Microbiol 48 549 559
6. WangG
AlamuriP
HumayunMZ
TaylorDE
MaierRJ
2005 The Helicobacter pylori MutS protein confers protection from oxidative DNA damage. Mol Microbiol 58 166 176
7. FriedbergEC
WalkerGC
SiedeW
WoodRD
SchultzRA
EllenbergerT
2005 DNA Repair and Mutagenesis Washington ASM Press 463
8. BuchmeierNA
LibbySJ
XuY
LoewenPC
SwitalaJ
1995 DNA repair is more important than catalase for Salmonella virulence in mice. J Clin Invest 95 1047 1053
9. CuccuiJ
EastonA
ChuKK
BancroftGJ
OystonPC
2007 Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect Immun 75 1186 1195
10. MertensK
LantsheerL
EnnisDG
SamuelJE
2008 Constitutive SOS expression and damage-inducible AddAB-mediated recombinational repair systems for Coxiella burnetii as potential adaptations for survival within macrophages. Mol Microbiol 69 1411 1426
11. SimmonsLA
FotiJJ
CohenSE
WalkerGC
2008 Escherichia coli and Salmonella: cellular and molecular biology;
NystromT
SlauchJM
SquiresCL
Washington, D. C. ASM Press
12. GiraudA
MaticI
TenaillonO
ClaraA
RadmanM
2001 Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291 2606 2608
13. LeClercJE
LiB
PayneWL
CebulaTA
1996 High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274 1208 1211
14. OliverA
CantonR
CampoP
BaqueroF
BlazquezJ
2000 High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288 1251 1254
15. MaticI
RadmanM
TaddeiF
PicardB
DoitC
1997 Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science 277 1833 1834
16. De GrooteMA
OchsnerUA
ShilohMU
NathanC
McCordJM
1997 Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc Natl Acad Sci U S A 94 13997 14001
17. MacMickingJD
NathanC
HomG
ChartrainN
FletcherDS
1995 Altered responses to bacterial infection and endotoxic shock in mice lacking inducible nitric oxide synthase. Cell 81 641 650
18. ImlayJA
2008 Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77 755 776
19. PooleRK
2005 Nitric oxide and nitrosative stress tolerance in bacteria. Biochem Soc Trans 33 176 180
20. BangIS
LiuL
Vazquez-TorresA
CrouchML
StamlerJS
2006 Maintenance of nitric oxide and redox homeostasis by the salmonella flavohemoglobin hmp. J Biol Chem 281 28039 28047
21. NelsonET
ClementsJD
FinkelsteinRA
1976 Vibrio cholerae adherence and colonization in experimental cholera: electron microscopic studies. Infect Immun 14 527 547
22. SchrankGD
VerweyWF
1976 Distribution of cholera organisms in experimental Vibrio cholerae infections: proposed mechanisms of pathogenesis and antibacterial immunity. Infect Immun 13 195 203
23. FordtranJS
WalshJH
1973 Gastric acid secretion rate and buffer content of the stomach after eating. Results in normal subjects and in patients with duodenal ulcer. J Clin Invest 52 645 657
24. SmithJL
2003 The role of gastric acid in preventing foodborne disease and how bacteria overcome acid conditions. J Food Prot 66 1292 1303
25. DuncanC
DougallH
JohnstonP
GreenS
BroganR
1995 Chemical generation of nitric oxide in the mouth from the enterosalivary circulation of dietary nitrate. Nat Med 1 546 551
26. LundbergJO
WeitzbergE
GladwinMT
2008 The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat Rev Drug Discov 7 156 167
27. ManningPB
CoulterST
JennessR
1968 Determination of nitrate and nitrite in milk and dry milk products. J Dairy Sci 51 1725 1730
28. DykhuizenRS
FrazerR
DuncanC
SmithCC
GoldenM
1996 Antimicrobial effect of acidified nitrite on gut pathogens: importance of dietary nitrate in host defense. Antimicrob Agents Chemother 40 1422 1425
29. IovineNM
PursnaniS
VoldmanA
WassermanG
BlaserMJ
2008 Reactive nitrogen species contribute to innate host defense against Campylobacter jejuni. Infect Immun 76 986 993
30. RaoA
JumpRL
PultzNJ
PultzMJ
DonskeyCJ
2006 In vitro killing of nosocomial pathogens by acid and acidified nitrite. Antimicrob Agents Chemother 50 3901 3904
31. XuJ
XuX
VerstraeteW
2001 The bactericidal effect and chemical reactions of acidified nitrite under conditions simulating the stomach. J Appl Microbiol 90 523 529
32. BourretTJ
PorwollikS
McClellandM
ZhaoR
GrecoT
2008 Nitric oxide antagonizes the acid tolerance response that protects Salmonella against innate gastric defenses. PLoS One 3 e1833
33. LijinskyW
1977 Nitrosamines and nitrosamides in the etiology of gastrointestinal cancer. Cancer 40 2446 2449
34. CrawfordMJ
GoldbergDE
1998 Role for the Salmonella flavohemoglobin in protection from nitric oxide. J Biol Chem 273 12543 12547
35. KimCC
MonackD
FalkowS
2003 Modulation of virulence by two acidified nitrite-responsive loci of Salmonella enterica serovar Typhimurium. Infect Immun 71 3196 3205
36. VenkateshJ
KumarP
KrishnaPS
ManjunathR
VarshneyU
2003 Importance of uracil DNA glycosylase in Pseudomonas aeruginosa and Mycobacterium smegmatis, G+C-rich bacteria, in mutation prevention, tolerance to acidified nitrite, and endurance in mouse macrophages. J Biol Chem 278 24350 24358
37. MullerCA
AutenriethIB
PeschelA
2005 Innate defenses of the intestinal epithelial barrier. Cell Mol Life Sci 62 1297 1307
38. QuinonesM
DavisBM
WaldorMK
2006 Activation of the Vibrio cholerae SOS response is not required for intestinal cholera toxin production or colonization. Infect Immun 74 927 930
39. LjungquistS
LindahlT
Howard-FlandersP
1976 Methyl methane sulfonate-sensitive mutant of Escherichia coli deficient in an endonuclease specific for apurinic sites in deoxyribonucleic acid. J Bacteriol 126 646 653
40. YajkoDM
WeissB
1975 Mutations simultaneously affecting endonuclease II and exonuclease III in Escherichia coli. Proc Natl Acad Sci U S A 72 688 692
41. CunninghamRP
SaporitoSM
SpitzerSG
WeissB
1986 Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol 168 1120 1127
42. CoxEC
DegnenGE
ScheppeML
1972 Mutator gene studies in Escherichia coli: the mutS gene. Genetics 72 551 567
43. DempleB
HalbrookJ
LinnS
1983 Escherichia coli xth mutants are hypersensitive to hydrogen peroxide. J Bacteriol 153 1079 1082
44. BelfortM
Pedersen-LaneJ
1984 Genetic system for analyzing Escherichia coli thymidylate synthase. J Bacteriol 160 371 378
45. DutraBE
LovettST
2006 Cis and trans-acting effects on a mutational hotspot involving a replication template switch. J Mol Biol 356 300 311
46. SimmonsLA
DaviesBW
GrossmanAD
WalkerGC
2008 Beta clamp directs localization of mismatch repair in Bacillus subtilis. Mol Cell 29 291 301
47. VasudevanSG
ArmaregoWL
ShawDC
LilleyPE
DixonNE
1991 Isolation and nucleotide sequence of the hmp gene that encodes a haemoglobin-like protein in Escherichia coli K-12. Mol Gen Genet 226 49 58
48. GardnerAM
GardnerPR
2002 Flavohemoglobin detoxifies nitric oxide in aerobic, but not anaerobic, Escherichia coli. Evidence for a novel inducible anaerobic nitric oxide-scavenging activity. J Biol Chem 277 8166 8171
49. AbbasK
BretonJ
PicotCR
QuesniauxV
BoutonC
2009 Signaling events leading to peroxiredoxin 5 up-regulation in immunostimulated macrophages. Free Radic Biol Med 47 794 802
50. FangFC
DeGrooteMA
FosterJW
BaumlerAJ
OchsnerU
1999 Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc Natl Acad Sci U S A 96 7502 7507
51. HebrardM
VialaJP
MeresseS
BarrasF
AusselL
2009 Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J Bacteriol 191 4605 4614
52. SigaudS
BecquetV
FrendoP
PuppoA
HerouartD
1999 Differential regulation of two divergent Sinorhizobium meliloti genes for HPII-like catalases during free-living growth and protective role of both catalases during symbiosis. J Bacteriol 181 2634 2639
53. Vazquez-TorresA
StevaninT
Jones-CarsonJ
CastorM
ReadRC
2008 Analysis of nitric oxide-dependent antimicrobial actions in macrophages and mice. Methods Enzymol 437 521 538
54. MerrellDS
CamilliA
1999 The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol Microbiol 34 836 849
55. PaulyGT
HughesSH
MoschelRC
1998 Comparison of mutagenesis by O6-methyl - and O6-ethylguanine and O4-methylthymine in Escherichia coli using double-stranded and gapped plasmids. Carcinogenesis 19 457 461
56. GabrielC
CaminsA
SuredaFX
AquirreL
EscubedoE
1997 Determination of nitric oxide generation in mammalian neurons using dichlorofluorescin diacetate and flow cytometry. J Pharmacol Toxicol Methods 38 93 98
57. KestonAS
BrandtR
1965 The Fluorometric Analysis of Ultramicro Quantities of Hydrogen Peroxide. Anal Biochem 11 1 5
58. PosselH
NoackH
AugustinW
KeilhoffG
WolfG
1997 2,7-Dihydrodichlorofluorescein diacetate as a fluorescent marker for peroxynitrite formation. FEBS Lett 416 175 178
59. AngelichioMJ
SpectorJ
WaldorMK
CamilliA
1999 Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect Immun 67 3733 3739
60. QadriF
ChowdhuryMI
FaruqueSM
SalamMA
AhmedT
2007 Peru-15, a live attenuated oral cholera vaccine, is safe and immunogenic in Bangladeshi toddlers and infants. Vaccine 25 231 238
61. RitchieJM
RuiH
BronsonRT
WaldorMK
2010 Back to the future: studying cholera pathogenesis using infant rabbits. MBio 1 e00047 00010
62. MerrellDS
HavaDL
CamilliA
2002 Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol Microbiol 43 1471 1491
63. AlvarezB
Ferrer-SuetaG
RadiR
1998 Slowing of peroxynitrite decomposition in the presence of mannitol and ethanol. Free Radic Biol Med 24 1331 1337
64. AbuaitaBH
WitheyJH
2009 Bicarbonate Induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect Immun 77 4111 4120
65. IdeH
TedzukaK
ShimzuH
KimuraY
PurmalAA
1994 Alpha-deoxyadenosine, a major anoxic radiolysis product of adenine in DNA, is a substrate for Escherichia coli endonuclease IV. Biochemistry 33 7842 7847
66. GeeJM
ValderasMW
KovachME
GrippeVK
RobertsonGT
2005 The Brucella abortus Cu,Zn superoxide dismutase is required for optimal resistance to oxidative killing by murine macrophages and wild-type virulence in experimentally infected mice. Infect Immun 73 2873 2880
67. MillerVL
MekalanosJJ
1988 A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170 2575 2583
68. MetcalfWW
JiangW
DanielsLL
KimSK
HaldimannA
1996 Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35 1 13
69. BaselskiVS
ParkerCD
1978 Intestinal distribution of Vibrio cholerae in orally infected infant mice: kinetics of recovery of radiolabel and viable cells. Infect Immun 21 518 525
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Compensatory Evolution of Mutations Restores the Fitness Cost Imposed by β-Lactam Resistance inČlánek The C-Terminal Domain of the Arabinosyltransferase EmbC Is a Lectin-Like Carbohydrate Binding Module
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 2- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- A Fresh Look at the Origin of , the Most Malignant Malaria Agent
- In Situ Photodegradation of Incorporated Polyanion Does Not Alter Prion Infectivity
- Highly Efficient Protein Misfolding Cyclic Amplification
- Positive Signature-Tagged Mutagenesis in : Tracking Patho-Adaptive Mutations Promoting Airways Chronic Infection
- Charge-Surrounded Pockets and Electrostatic Interactions with Small Ions Modulate the Activity of Retroviral Fusion Proteins
- Whole-Body Analysis of a Viral Infection: Vascular Endothelium is a Primary Target of Infectious Hematopoietic Necrosis Virus in Zebrafish Larvae
- Inhibition of Nox2 Oxidase Activity Ameliorates Influenza A Virus-Induced Lung Inflammation
- STAT2 Mediates Innate Immunity to Dengue Virus in the Absence of STAT1 via the Type I Interferon Receptor
- Uropathogenic P and Type 1 Fimbriae Act in Synergy in a Living Host to Facilitate Renal Colonization Leading to Nephron Obstruction
- Elite Suppressors Harbor Low Levels of Integrated HIV DNA and High Levels of 2-LTR Circular HIV DNA Compared to HIV+ Patients On and Off HAART
- DC-SIGN Mediated Sphingomyelinase-Activation and Ceramide Generation Is Essential for Enhancement of Viral Uptake in Dendritic Cells
- Short-Lived IFN-γ Effector Responses, but Long-Lived IL-10 Memory Responses, to Malaria in an Area of Low Malaria Endemicity
- Induces T-Cell Lymphoma and Systemic Inflammation
- The C-Terminus of RON2 Provides the Crucial Link between AMA1 and the Host-Associated Invasion Complex
- Critical Role of the Virus-Encoded MicroRNA-155 Ortholog in the Induction of Marek's Disease Lymphomas
- Type I Interferon Signaling Regulates Ly6C Monocytes and Neutrophils during Acute Viral Pneumonia in Mice
- Atypical/Nor98 Scrapie Infectivity in Sheep Peripheral Tissues
- Innate Sensing of HIV-Infected Cells
- BosR (BB0647) Controls the RpoN-RpoS Regulatory Pathway and Virulence Expression in by a Novel DNA-Binding Mechanism
- Compensatory Evolution of Mutations Restores the Fitness Cost Imposed by β-Lactam Resistance in
- Expression of Genes Involves Exchange of the Histone Variant H2A.Z at the Promoter
- The RON2-AMA1 Interaction is a Critical Step in Moving Junction-Dependent Invasion by Apicomplexan Parasites
- Epstein-Barr Virus Nuclear Antigen 3C Facilitates G1-S Transition by Stabilizing and Enhancing the Function of Cyclin D1
- Transcription and Translation Products of the Cytolysin Gene on the Mobile Genetic Element SCC Regulate Virulence
- Phosphatidylinositol 3-Monophosphate Is Involved in Apicoplast Biogenesis
- The Rubella Virus Capsid Is an Anti-Apoptotic Protein that Attenuates the Pore-Forming Ability of Bax
- Episomal Viral cDNAs Identify a Reservoir That Fuels Viral Rebound after Treatment Interruption and That Contributes to Treatment Failure
- Genetic Mapping Identifies Novel Highly Protective Antigens for an Apicomplexan Parasite
- Relationship between Functional Profile of HIV-1 Specific CD8 T Cells and Epitope Variability with the Selection of Escape Mutants in Acute HIV-1 Infection
- The Genotype of Early-Transmitting HIV gp120s Promotes αβ –Reactivity, Revealing αβ/CD4 T cells As Key Targets in Mucosal Transmission
- Small Molecule Inhibitors of RnpA Alter Cellular mRNA Turnover, Exhibit Antimicrobial Activity, and Attenuate Pathogenesis
- The bZIP Transcription Factor MoAP1 Mediates the Oxidative Stress Response and Is Critical for Pathogenicity of the Rice Blast Fungus
- Entrapment of Viral Capsids in Nuclear PML Cages Is an Intrinsic Antiviral Host Defense against Varicella-Zoster Virus
- NS2 Protein of Hepatitis C Virus Interacts with Structural and Non-Structural Proteins towards Virus Assembly
- Measles Outbreak in Africa—Is There a Link to the HIV-1 Epidemic?
- New Models of Microsporidiosis: Infections in Zebrafish, , and Honey Bee
- The C-Terminal Domain of the Arabinosyltransferase EmbC Is a Lectin-Like Carbohydrate Binding Module
- A Viral microRNA Cluster Strongly Potentiates the Transforming Properties of a Human Herpesvirus
- Infections in Cells: Transcriptomic Characterization of a Novel Host-Symbiont Interaction
- Secreted Bacterial Effectors That Inhibit Host Protein Synthesis Are Critical for Induction of the Innate Immune Response to Virulent
- Genital Tract Sequestration of SIV following Acute Infection
- Functional Coupling between HIV-1 Integrase and the SWI/SNF Chromatin Remodeling Complex for Efficient Integration into Stable Nucleosomes
- DNA Damage and Reactive Nitrogen Species are Barriers to Colonization of the Infant Mouse Intestine
- The ESCRT-0 Component HRS is Required for HIV-1 Vpu-Mediated BST-2/Tetherin Down-Regulation
- Targeted Disruption of : Invasion of Erythrocytes by Using an Alternative Py235 Erythrocyte Binding Protein
- Trivalent Adenovirus Type 5 HIV Recombinant Vaccine Primes for Modest Cytotoxic Capacity That Is Greatest in Humans with Protective HLA Class I Alleles
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genetic Mapping Identifies Novel Highly Protective Antigens for an Apicomplexan Parasite
- Type I Interferon Signaling Regulates Ly6C Monocytes and Neutrophils during Acute Viral Pneumonia in Mice
- Infections in Cells: Transcriptomic Characterization of a Novel Host-Symbiont Interaction
- The ESCRT-0 Component HRS is Required for HIV-1 Vpu-Mediated BST-2/Tetherin Down-Regulation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



