-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaRSV-Induced H3K4 Demethylase KDM5B Leads to Regulation of Dendritic Cell-Derived Innate Cytokines and Exacerbates Pathogenesis
Respiratory syncytial virus (RSV) is a significant public health concern. Nearly all children are infected by two years of age, and severe infection often results in hospitalization. There is no vaccine for RSV, and previous attempts have resulted in increased disease severity in immunized children once they were exposed to the virus. Therefore, a better understanding of how RSV directs the immune response is needed. In this study, we found that the protein KDM5B regulates an epigenetic mechanism that directs the immune response to RSV. KDM5B suppressed the activation of key antiviral signals in dendritic cells, and inhibition of KDM5B led to gene activation and increased antiviral function. This correlated with decreased pathology in the lungs. Therefore, our data suggest that new attempts at designing a vaccine should consider the effects of vaccination on dendritic cells, and should consider strategies that will increase antiviral signals from dendritic cells.
Published in the journal: . PLoS Pathog 11(6): e32767. doi:10.1371/journal.ppat.1004978
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004978Summary
Respiratory syncytial virus (RSV) is a significant public health concern. Nearly all children are infected by two years of age, and severe infection often results in hospitalization. There is no vaccine for RSV, and previous attempts have resulted in increased disease severity in immunized children once they were exposed to the virus. Therefore, a better understanding of how RSV directs the immune response is needed. In this study, we found that the protein KDM5B regulates an epigenetic mechanism that directs the immune response to RSV. KDM5B suppressed the activation of key antiviral signals in dendritic cells, and inhibition of KDM5B led to gene activation and increased antiviral function. This correlated with decreased pathology in the lungs. Therefore, our data suggest that new attempts at designing a vaccine should consider the effects of vaccination on dendritic cells, and should consider strategies that will increase antiviral signals from dendritic cells.
Introduction
Respiratory syncytial virus (RSV) is a significant burden to healthcare worldwide. In the United States nearly all children have been infected by age two [1,2]. Severe infections are the leading cause of bronchiolitis in children, resulting in up to 125,000 hospitalizations in the US each year [3,4]. Furthermore, infants who are hospitalized with severe disease are 3–4 times more likely to develop asthma later in life [5,6], as RSV interferes with the initiation of the adaptive immune response leading to an altered immune environment in the lungs. One consequence is the recruitment of T cells that produce interleukin (IL)-4, IL-5 and IL-13, which are cytokines involved in the pathogenesis of lung disease, including asthma. Since RSV infection does not result in a strong memory response, repeated infections are common throughout childhood and for adults [7]. Currently, there is no vaccine for RSV. Attempts to create a formalin-inactivated vaccine resulted in more severe infection upon exposure to the virus in those children who were vaccinated compared to unvaccinated children [8]. Thus, the need to understand the immune response to RSV at the molecular level is critical to develop better therapeutics and aid in the development of effective vaccines.
Dendritic cells (DCs) residing in the airway are among the first cells to encounter an RSV infection. An important role for DCs is the production of IL-12, which is critical to initiate a Th1 response following infection with a virus. Viral infection also results in the recognition of pathogen-associated molecular patterns (PAMPs) such as viral nucleic acids, including double-stranded RNA. DCs express toll-like receptors (TLRs) that recognize these PAMPs. Viral RNA recognition signals induce the production of type I IFNs, including IFN-β, thereby leading to an antiviral state in nearby cells and initiating the proper T cell response. However, while other respiratory viruses such as influenza drive a strong IFN-β response, RSV infection suppresses this response, which is partially due to viral non-structural protein interactions with activation factors, resulting in the suppression of both helicase RIG-I [9] and the transcription factor NF-γB [10]. These events not only allow for viral replication, but also promote an altered immune environment in the lungs, characterized by mucus hypersecretion [11]. As DCs are important cells in directing the T cell response, it is imperative to understand the mechanisms by which RSV interferes with proper DC function.
Recent studies have documented that epigenetic modifications of immune cells at the chromatin level contribute to either activation or repression of specific genes. This regulation can occur by several mechanisms, including methylation, acetylation and phosphorylation of histone tails. Histone methylation can occur on lysine and arginine residues, and contribute to activation or inhibition of transcription. For example, the addition of methyl groups on lysine (K) 27 of histone (H) 3 (i.e. H3K27) is associated with repression of gene transcription, whereas H3K4 methylation correlates with active gene transcription [12–14]. This process is controlled by methyltransferases that add methyl groups and demethylases that remove them. There is accumulating evidence that epigenetic modulation is an important component of immune cell phenotype and function. For example, the cytokines typically produced by Th2 cells, including IL-4, IL-5 and IL-13, are silenced in Th1 cells through histone modification [15]. In addition, Th17 cells have increased H3K4 methylation, a gene activating mark, at the Th17 promoter [16]. In DCs, IL-12 production is decreased following severe sepsis due to decreased H3K4 methylation altering subsequent immune responses [17]. Despite these inroads, data is lacking regarding the contribution of epigenetic regulation to immune cell activation. In the present studies an upregulation of Kdm5b, coding for an H3K4 demethylase, following RSV infection of DCs was observed. As H3K4 methylation is an activating mark, this demethylase has the potential to repress transcription. A role for KDM5B in transcriptional repression has been reported in cancer cells including melanoma, breast cancer and prostate cancer [18–20]. Here we show that KDM5B has a critical role in preventing the activation of DCs during RSV-induced immune responses. Our results show that decreasing Kdm5b expression by siRNA, chemical inhibition or genetic deletion prior to RSV infection leads to an increase in the production of IFN-β and other inflammatory cytokines compared to uninfected controls, as well as decreased Th2 pathogenesis in vivo thus linking Kdm5b expression with disease exacerbation during RSV infection.
Results
Expression of histone lysine demethylases and Kdm5b following infection of BMDCs with RSV
A previous report has identified a role for epigenetic regulation in immune cells following viral infection [21]. As DCs are critical for priming the T cell response to RSV infection, studies were initiated to determine whether exposing DCs to RSV resulted in changes in the expression of epigenetic factors in the DCs. BMDCs were infected with RSV or activated by p(I:C) or imiquimod, the ligands for TLR3 and TLR7 respectively, as RSV is known to activate cells through both TLR3 and TLR7 [22,23], in addition to other mechanisms. In order to observe early gene expression of epigenetic enzymes, RNA was harvested at 4 hours post treatment to examine transcription levels of genes coding for “epigenetic” enzymes by qPCR array. Several classes of enzymes were analyzed including histone deacetylases (HDACs), histone lysine demethylases (KDMs), protein arginine methyltransferases (PRMTs), and histone lysine methyltransferases (KMTs) (Fig 1A). A defining observation was the upregulation of Kdm5b demethylase by RSV in contrast to the downregulation of this enzyme by stimulation through TLR3 and TLR7 (Fig 1A). While Kdm6b was upregulated by RSV infection of DCs, this enzyme was also significantly upregulated by treatment of cells with imiquimod. Because Kdm5b was upregulated only by RSV, studies focused on Kdm5b as a potential unique enzyme in the DC response to RSV. PCR analysis confirmed the peak expression of Kdm5b in BMDCs at 12 hours following RSV infection (Fig 1B). Furthermore, while Kdm5b was upregulated in BMDCs infected with RSV, it was not upregulated by influenza (H1N1) virus, nor in RSV-infected epithelial cells or alveolar macrophages (S1 Fig). Therefore, studies focused on H3K4 demethylase Kdm5b and its role on perturbing critical innate immune genes in DCs.
Fig. 1. Kdm5b expression increases following infection of BMDCs with RSV. 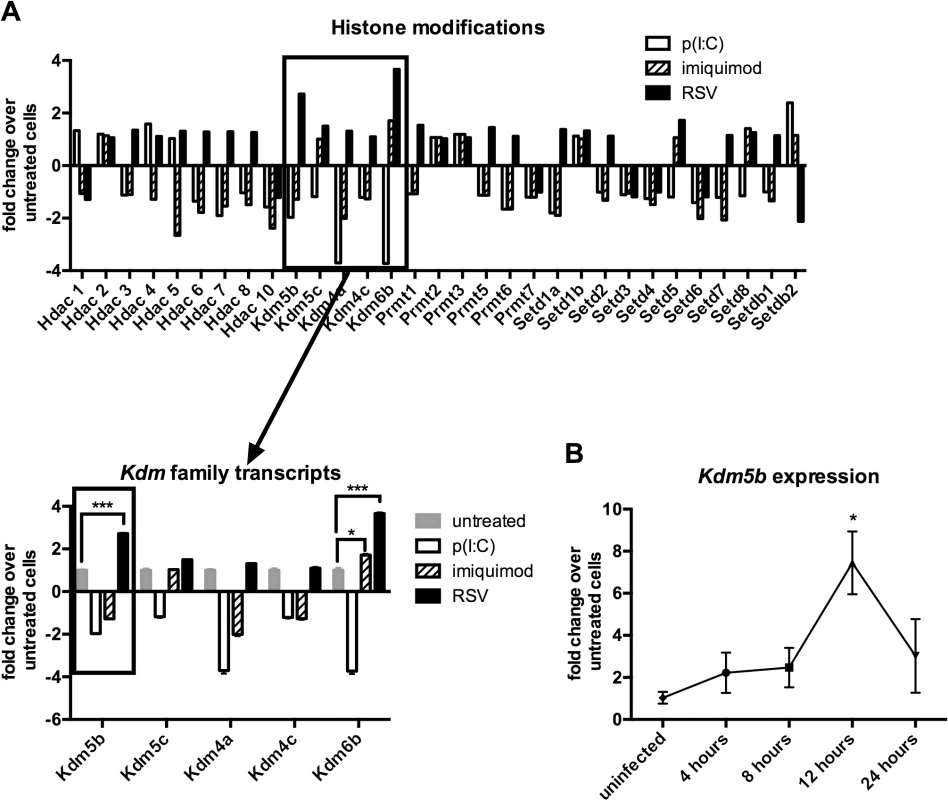
Bone marrow-derived dendritic cells were infected with RSV (MOI = 1) or treated with poly(I:C) (20 μg/ml) or imiquimod (1 μg/ml) for four hours. RNA was extracted and reverse transcribed, and the cDNA was used in a PCR array to measure changes in the expression of epigenetic enzymes (A), including the histone lysine demethylase family. Expression of Kdm5b was measured over a time course following infection with RSV (B). n = 3–5 samples/group, and data are representative of three independent experiments. *p<0.05. Increased expression of pro-inflammatory cytokines following disruption of KDM5B function
To determine whether KDM5B affects DC function, specific siRNA was used to knock down Kdm5b resulting in >70% reduction in expression levels (Fig 2A). Previous reports have indicated that RSV, unlike many viruses, is a poor inducer of type I IFN, including IFN-β [9,10]. BMDCs infected with RSV produced low levels of IFN-β at both 4 and 24 hours, whereas H1N1 virus produced very high levels (S2 Fig). We therefore hypothesized that the increase in KDM5B in BMDCs contributed to the suppression of type I IFN production and that knocking down Kdm5b expression would result in increased IFN-β. Following in vitro treatment of BMDCs with Kdm5b-specific siRNA or with a scrambled siRNA control, significantly increased expression levels of Ifnb, as well as the pro-inflammatory cytokines Tnfa and Il6 were observed in in vitro RSV-infected cells compared to sham-infected BMDCs (Fig 2B). To determine whether APC function was affected by Kdm5b siRNA or inhibitor treatment, MHC-II expression on the cell surface of BMDCs was measured, as well as expression of the co-stimulatory molecules CD80 and CD86. No differences in any maturation markers were noticed in treated cells compared to controls (S3 Fig). Furthermore, when a chemical inhibitor, 2,4-pyridinedicarboxylic acid (2,4-PDCA), was used to block the function of KDM5B [24,25] prior to RSV infection, significantly higher levels of Ifnb, Tnfa and Il6 transcripts compared to controls were observed (Fig 2C). While this inhibitor also interacts with other KDM family members, it has the highest specificity for KDM5B. Thus, two independent approaches to block KDM5B function demonstrated an altered immune response resulting in increases of critical innate cytokines.
Fig. 2. siRNA knockdown of Kdm5b leads to increased cytokine and chemokine gene expression. 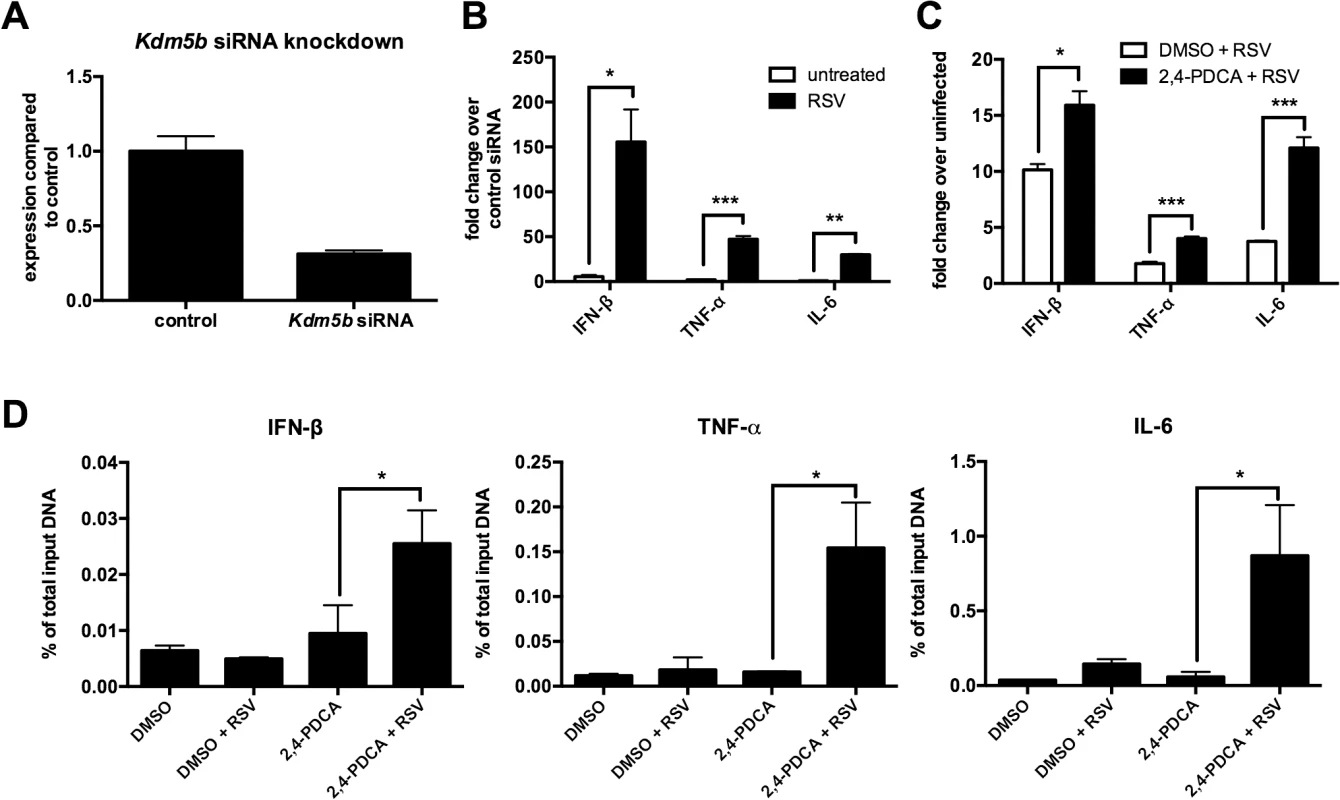
(A) BMDCs were transfected with Kdm5b-specific siRNA or non-targeting control siRNA (1 μM). Total RNA was extracted from the cells and gene expression levels measured by qPCR. n = 6 from two combined experiments. (B) Transcript levels of Ifnb, Tnf and Il6 were measured from Kdm5b-siRNA transfected BMDCs and were compared to scrambled siRNA control following infection with RSV. n = 3 samples/group and is representative of two independent experiments. (C) BMDCs were treated with 1μM 2,4-PDCA or 0.1% DMSO for 24 hours and subsequently infected with RSV for 24 hours. Transcript levels were measured by qPCR. n = 4 samples/group and is representative of two independent experiments. (D) DMSO or 2,4-PDCA-treated BMDCs were infected with RSV for 24 hours. A ChIP assay was performed to determine the H3K4me3 status at the promoter regions of the Ifnb, Tnf and Il6 genes. Data are representative of three different experiments, with each sample run in duplicate. n = 4 samples/group. *p<0.05, **p<0.01, ***p<0.001. KDM5B catalyzes the demethylation of H3K4me3 and H3K4me2. As H3K4me3 is associated with active gene transcription, the activity of KDM5B in removing a methyl group leads to decreased promoter activity and decreased gene transcription. Since blocking KDM5B activity led to increased levels of proinflammatory cytokines, we hypothesized that blocking the demethylase activity would lead to greater H3K4me3 at the promoters of these cytokines. To test this hypothesis, a ChIP assay using an anti-H3K4me3 antibody was performed on cells treated with 2,4-PDCA, and primers designed to recognize the promoter regions of Ifnb, Tnf and Il6 were used. Treatment of DC with 2,4-PDCA prior to infection with RSV led to an increase of H3K4me3 compared to controls on all three cytokine promoters (Fig 2D). Conversely, it was found that there were no differences in H3K4 methylation on the promoters of Il10 and Il12, which are two cytokines that were unchanged following RSV infection of Kdm5b-deficient DCs (S4 Fig). These results indicate that the KDM5B demethylase activity acts on the promoters of specific inflammatory genes, thereby suppressing transcription.
Inhibition of KDM5B in human DCs results in decreased inflammatory gene activity and lower IL-4 production by co-cultured CD4+ T cells
The above data show that inhibiting KDM5B function leads to increased innate cytokine production after RSV infection. To determine the role of KDM5B in human cells, human monocyte-derived DCs (MoDCs) cultured from peripheral blood monocytes were used. As shown in Fig 3A, KDM5B is significantly upregulated in MoDCs at 12 and 24 hours following RSV infection, and although the degree of upregulation is less pronounced than in mouse DCs, the kinetics are very similar (Fig 3A). To assess the role of KDM5B in human DCs, the function of KDM5B was inhibited by treating the cells with 2,4-PDCA for 24 hours, followed by RSV infection. Similar to observations in mouse cells treated with 2,4-PDCA, inhibiting KDM5B in human MoDCs led to increased production of IFNB, TNF and IL6 compared to RSV alone or DMSO control (Fig 3B). To determine whether the presence of the inhibitor affected the H3K4me3 status of these genes, a ChIP analysis for H3K4 was performed and examined the promoter regions of specific innate cytokine genes. Interestingly, MoDCs exhibited a slight increase in promoter methylation following incubation with the inhibitor alone, which was further increased when the cells were infected with RSV (Fig 3C). Similar to observations in mouse DCs, no change in methylation at the IL10 an IL12 promoters was observed (S4 Fig). Finally, infected MoDCs that had been treated with DMSO or 2,4-PDCA were cultured with autologous CD4+ T cells in the presence of RSV to assess the APC function of the MoDCs (Fig 3D). While the T cells co-cultured with 2,4-PDCA-treated DCs had similar levels of IFN-γ production, the Th2 cytokines IL-5 and IL-13 were significantly decreased. These data suggest that inhibiting KDM5B function in human MoDCs results in regulation of Th2 cytokine production, supporting the hypothesis that RSV drives an altered immune phenotype that relies on epigenetic regulation of DC.
Fig. 3. Inhibiting KDM5B in human monocyte-derived DCs leads to increased cytokine production. 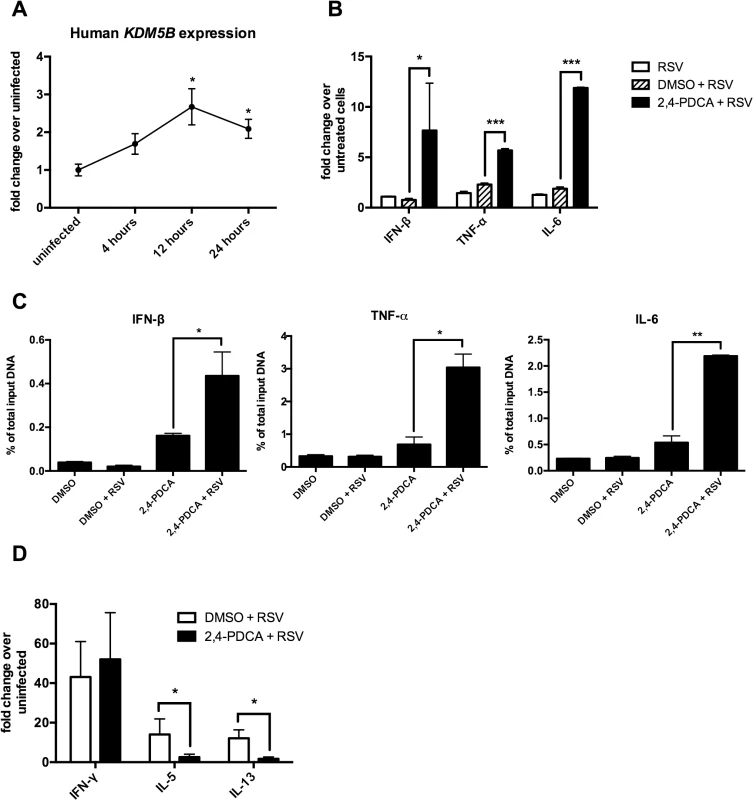
(A) DCs were grown from blood monocytes and infected with RSV. RNA was isolated and KDM5B expression was determined by qPCR. n = 6 samples from three pooled donors. (B) DCs were treated with DMSO or 2,4-PDCA for 24 hours, and were then infected with RSV for 24 hours. Cytokine levels were measured by qPCR. n = 3 samples/group and is representative of three independent experiments. (C) DMSO or 2,4-PDCA-treated BMDCs were infected with RSV for 24 hours. A ChIP assay was performed to determine the H3K4me3 status at the promoter regions of the IFNB, TNF and IL6 genes. n = 4 samples/group. (D) DCs were treated with DMSO or 2,4-PDCA and infected with RSV. Autologous CD4+ T cells were cultured with the DCs for 48 hours. mRNA was extracted and cytokines levels measured by qPCR. n = 3 samples/group and data are representative of two independent experiments. p<0.05,**p<0.01 ***p<0.001. Transfer of RSV-infected Kdm5b-deficient DCs leads to a decreased pathogenic pulmonary RSV challenge
Pro-inflammatory cytokines, including IFN-β, are important factors in driving Th1 responses following viral infection in vivo [26]. The above data indicated siRNA knockdown of Kdm5b led to increased pro-inflammatory cytokines in vitro, suggesting that priming the immune system in vivo with these DCs would drive a stronger Th1 response to RSV. To test this hypothesis, BMDCs were treated with Kdm5b-specific siRNA or scrambled control siRNA for 48 hours and subsequently infected overnight with RSV. These cells were then transferred intratracheally into naïve C57Bl/6 mice to prime the immune system in the context of Kdm5b-deficient DCs. One week after transfer, mice were challenged with RSV (Fig 4A). Our lab has previously demonstrated that this sensitization protocol with myeloid DC elicits a pathogenic immune environment upon RSV reinfection, and that the RSV-infected DCs that are transferred begin migrating to the lymph nodes within 24 hours [27,28]. Eight days following challenge, the mediastinal lymph nodes (MLN) were removed and restimulated ex vivo to determine the effect that sensitizing mice with Kdm5b-knockdown DCs would have on the T cell response. The data indicated an increase in IFN-γ production from mice that had been sensitized with Kdm5b-specific siRNA treated DCs, accompanied by a significant decrease in IL-5 and a trend toward less IL-4 production (Fig 4B). Furthermore, it was found that the lungs of mice primed with DCs that had been transfected with Kdm5b-specific siRNA had decreased inflammatory infiltrates compared to mice primed with control siRNA-treated DCs, as observed by H&E stained sections (Fig 4C). High-power images revealed that the inflammation surrounding the airways of control RSV-infected DC-sensitized mice contained numerous eosinophils (Fig 4C, insets). The number of eosinophils surrounding the airways was enumerated by microscopy in 100μm sections demonstrating decreased numbers in the Kdm5b siRNA group (Fig 4D) and confirmed with flow cytometric analysis of Gr-1+ and Siglec-F+ cells (Fig 4E). Additionally, it was noted that the total number of cells in the lungs was increased in mice sensitized with control siRNA, but the lungs had fewer cells when the mice received Kdm5b siRNA (Fig 4F). Flow cytometric analysis showed that these differences in total lung cells were due to fewer numbers of both conventional CD11c+CD11b+ DCs, as well as CD11c+CD103+ DCs (Fig 4G) [29,30]. Furthermore, there was a decrease in total CD4+ T cells as well as activated CD4+CD69+ cells (Fig 4H). Increased numbers of total cells, as well as DCs, T cells and eosinophils were also observed in mice that where sensitized with uninfected DCs, but to a lesser degree. (S5 Fig). In these mice, the numbers of inflammatory cells were similar to the numbers of cells measured in mice that received Kdm5b-deficient DCs, but was lower than the number of cells noted with control-treated, RSV-infected DCs were used to prime mice prior to challenge. Thus, the inhibition of Kdm5b in RSV-infected DCs led to significant protection from immunopathology that often has been associated with immunization to RSV [8].
Fig. 4. Transfer of RSV-infected Kdm5b-deficient DCs leads to a decreased pathogenic pulmonary RSV challenge. 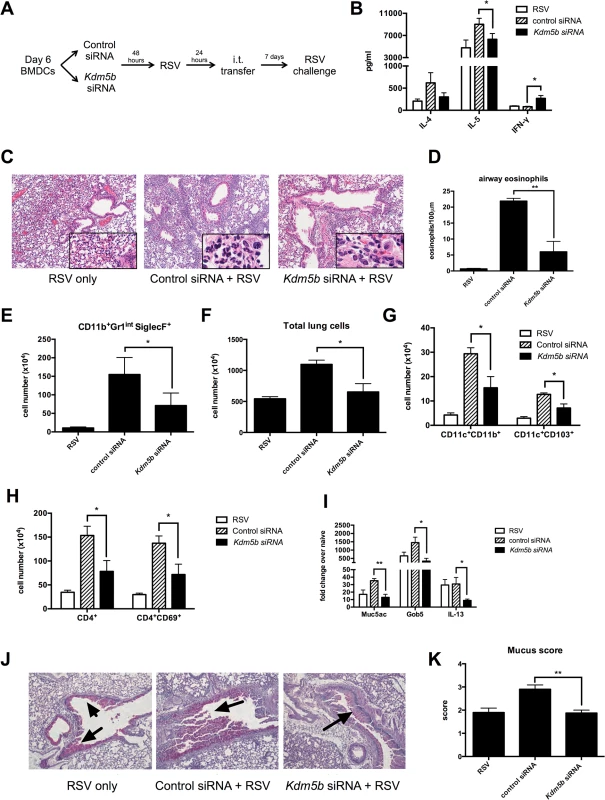
(A) BMDCs were transfected with Kdm5b-specific siRNA or non-targeting control, then infected with RSV overnight. These BMDCs were transferred intratracheally to the lungs of naïve mice 7 days prior to RSV infection. (B) At 8 days post infection, protein levels were measured from MLN cells restimulated with RSV in vitro. (C) Inflammation around the airways was measured by H&E staining in mice primed with BMDCs, including airway eosinophils (inset). (D) Eosinophils were counted in 100μm sections around the airways. (E) Lungs were dispersed into a single cell suspension using collagenase A and DNase. Esoinophils were measured using flow cytometry. (F) Total lung cells were counted following digestion of lung tissue. CD11c+CD11b+ and CD11c+CD103+ DC populations were measured by flow cytometry (G), as well as CD4+ T cells and activated CD4+CD69+ cells (H). (I) RNA was extracted from lung tissue using Trizol reagent, and transcripts of Muc5ac, Gob5 and Il13 were measured by qPCR. (J) Mucus was visualized in the lungs using PAS staining. Arrows indicate mucus production. (K) Identifying numbers on histology slides were blinded and slides were scored on a scale of 1–4 for mucus production. For all data n = 4-5samples/group and data are representative of two independent experiments. *p<0.05, **p<0.01. Mucus production is a common occurrence during RSV infection, and is linked to the Th2 response to the virus. When mice were primed with Kdm5b-specific siRNA transfected DCs prior to RSV challenge, lower levels of the mucus-associated genes Muc5ac and Gob5 were observed in the lungs of mice primed with Kdm5b-specific siRNA (Fig 4I). In addition, a primary inducer of goblet cell metaplasia and mucus hypersecretion, Il13, was also reduced in mice primed by Kdm5b siRNA inhibited DC (Fig 4I). Furthermore, mucus levels in the airways decreased, as visualized by PAS staining (Fig 4J). Scoring of mucus production on a scale of 1–4 is quantified in Fig 4K and demonstrated a significant decrease in overall mucus production in the Kdm5b-inhibited DC transfer model. These results indicate that priming the immune response to RSV with DCs lacking Kdm5b results in a less pathogenic immune environment in vivo.
Infection of Kdm5bf/f-CD11c-Cre+ mice results in decreased Th2 cytokines and decreased mucus production following RSV infection
To provide further genetic evidence for the role of Kdm5b, a DC-specific Kdm5b knockout mouse was developed. Kdm5bf/f mice [31] were crossed with CD11c-Cre+/- mice to create Kdm5b knockout CD11c+ DCs. Initial evaluation of these mice demonstrate no baseline differences in the number of immune cells in the lungs and spleens, including CD3+CD4+ and CD3+CD8+ T cells, CD11c+CD11b+ DCs and CD11b+F4/80+ macrophages (S6 Fig). BMDCs from Kdm5bf/f-CD11c-Cre+ and control Kdm5bf/f-CD11c-Cre- mice that were grown had no identifiable growth or differentiation defects. BMDC from Kdm5bf/f-CD11c-Cre+ mice infected with RSV demonstrated increased expression levels of the innate cytokines Ifnb, Tnf and Il6 at 24 hours compared to Cre- controls (Fig 5A), suggesting that KDM5B acts to suppress cytokine expression in DCs following RSV infection. These latter differences were not due to infectivity as there were no differences in the ability to infect the DCs from the Cre+ mice (S7 Fig). These mice were then infected with RSV, and cytokine transcripts were measured in whole lung tissue early after infection. Innate cytokine transcripts were elevated in Cre+ mice over Cre- controls at both 2 and 4 days post-infection (dpi) (Fig 5B). To determine whether this corresponded to the demethylase activity of KDM5B at the promoter regions of these genes, a ChIP assay in Kdm5b-deficient BMDCs and Cre- controls probing for H3K4 methylation was performed. Similar to observations using chemical inhibition of KDM5B, the Kdm5b-deficient DC had increased H3K4me3 at the promoter regions of all three inflammatory genes examined (Fig 5C) as assessed by ChIP analyses, but not of Il10 and Il12 (S4 Fig). These results further support the notion that KDM5B demethylates H3K4 at specific innate cytokine promoters, thereby contributing to gene suppression.
Fig. 5. Deletion of Kdm5b in DCs leads to increased cytokine production and decreased Th2 pathology in vivo. 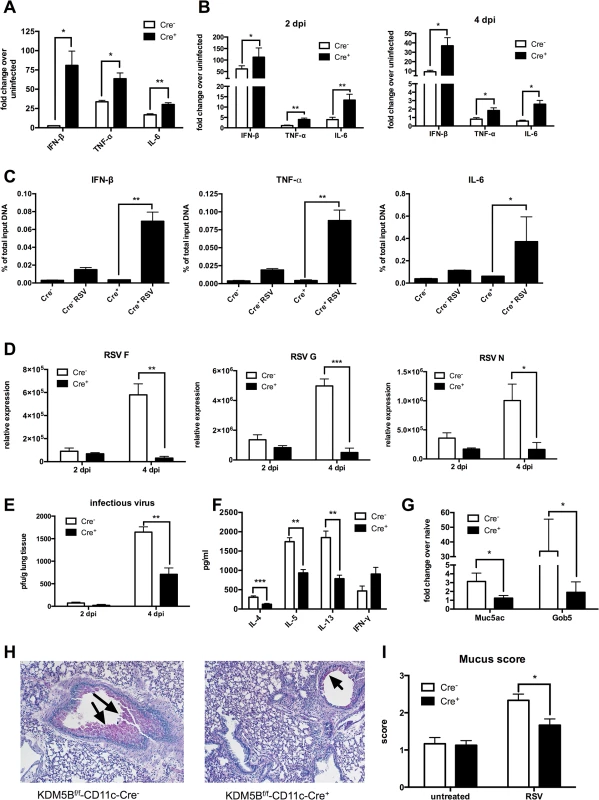
(A) DCs were cultured from bone marrow of Kdm5bf/f-CD11c-Cre+ mice and Cre- controls and were infected with RSV. RNA was extracted after 24 hours and cytokine production measured by qPCR. (B) Kdm5bf/f-CD11c-Cre+ mice and Cre- controls were infected intranasally with RSV and the lungs were removed at 2 and 4 dpi. Lung tissue was homogenized and RNA extracted. Cytokine levels were measured by qPCR, and are expressed as fold change over infected Cre- controls. (C) BMDCs were infected with RSV for 24 hours, and a ChIP assay was performed to determine the H3K4me3 status at the promoter regions of the Ifnb, Tnf and Il6 genes. (D) Lungs were removed from mice at 2 and 4 dpi and viral mRNA levels for the F, G and N genes were measured by qPCR. (E) Infectious virus was determined by plaque assay at 2 and 4 dpi. (F) MLN were removed from mice at 8 dpi and were restimulated in vitro with RSV for 48 hours. Proteins in the supernatant were measured by bioplex assay. (G) Mice were infected with RSV for eight days, then lungs were removed and homogenized, and RNA was extracted. Transcripts of Muc5ac and Gob5 were determined by qPCR. (H) Lung sections were stained with PAS to visualize mucus production at 8 dpi. Arrows indicate mucus production. (I) Mucus production was quantified from in lung sections on a scale of 1–4. *p<0.05, **p<0.01. The previous data above suggested a decreased Th2 response to RSV infection when mice were sensitized with Kdm5b-knockdown DCs. However, to determine whether KDM5B is important in primary RSV infection, Kdm5bf/f-CD11c-Cre+ and control mice were infected with RSV. At 2 and 4 dpi, lungs were removed and RNA was isolated from homogenates. mRNA levels of the RSV proteins F, G and N were measured (Fig 5D). Previous studies indicate that RSV replication peaks at 4 dpi [32]; no differences in RSV gene expression at 2 dpi were found, but by day 4, viral gene expression was significantly increased in Kdm5bf/f-CD11c-Cre- mice, whereas viral clearance was enhanced in Kdm5bf/f-CD11c-Cre+ mice. These results were consistent with a plaque assay, where there were no differences in the levels of infectious virus at 2 dpi, but at 4 dpi the Kdm5bf/f-CD11c-Cre+ mice had significantly fewer infectious particles in the lungs compared to controls (Fig 5E). At 8 dpi, MLN were removed and restimulated with RSV ex vivo. Similar to results shown in Fig 4, the data indicated that Th2 cytokines IL-4, IL-5 and IL-13 were substantially decreased in supernatants from restimulated cells, while IFN-γ production was increased, although not significantly (Fig 5F). Expression of the mucus-associated genes Muc5ac and Gob5 in the lung tissue of infected mice was also measured. Kdm5bf/f-CD11c-Cre+ mice had lower expression of mucus-associated genes compared to the control Cre- mice (Fig 5G). Visualization of mucus by histologic PAS staining demonstrated decreased mucus production in Kdm5bf/f-CD11c-Cre+ mice compared to controls (Fig 5H). Slides were scored on a scale of 1–4 for mucus production, and quantification showed less mucus production in Kdm5bf/f-CD11c-Cre+ mice (Fig 5I). Together, these data demonstrate that genetic deletion of Kdm5b from CD11c+ cells results in increased innate cytokine production by DCs and a correlative decrease in the Th2 response to RSV in vivo.
Sensitization with Kdm5bf/f-CD11c-Cre+ BMDCs prior to RSV infection leads to decreased inflammation, Th2 cytokine expression and mucus production in vivo
The above studies found a decrease in proinflammatory cytokines in Kdm5bf/f-CD11c-Cre+ BMDCs similar to that observed with siRNA-treated BMDCs. To confirm that our results in the Kdm5bf/f-CD11c-Cre+ mice were mediated by DCs, BMDCs were infected with RSV for 24 hours, then delivered intratracheally into mice. After 7 days, mice were infected with RSV and assessed at 8 dpi. We found that mice that received Kdm5bf/f-CD11c-Cre- DCs had increased inflammation compared to RSV only controls, but that this inflammation was decreased in mice that were sensitized with Kdm5bf/f-CD11c-Cre+ DCs, as determined by cellular infiltrates and H&E staining (Fig 6A). The MLN were removed and restimulated with RSV in vitro for 48 hours. Sensitizing mice with RSV-infected DCs led to increased levels of Th2 cytokines, IL-4, IL-5 and IL-13, but cytokine production was significantly lower when mice had been sensitized with Kdm5b/f-CD11c-Cre+ DCs compared to controls, although the decrease in IL-5 was not significant. These mice also had an increase, although not significant, in IFN-γ production from the lymph nodes (Fig 6B). Kdm5bf/f-CD11c-Cre+ DCs sensitized mice had fewer numbers of both CD11c+CD11b+ and CD11c+CD103+ DCs as well as fewer CD4+ T cells in the lungs. Also, fewer activated CD4+CD69+ T cells were observed in the lungs of Cre+ sensitized mice compared to mice sensitized with control DCs (Fig 6C and 6D). Finally, Kdm5bf/f-CD11c-Cre+ DC sensitized mice had less mucus production in the lungs compared to mice treated with control DCs, with a significant decrease in Gob5 expression (Fig 6E and 6F). Together, these results confirm the role of Kdm5b in skewing the immune environment towards increased pulmonary pathology.
Fig. 6. Sensitizing the lungs with Kdm5bf/f-CD11c-Cre+ DCs prior to infection results in decreased pathology following RSV challenge. 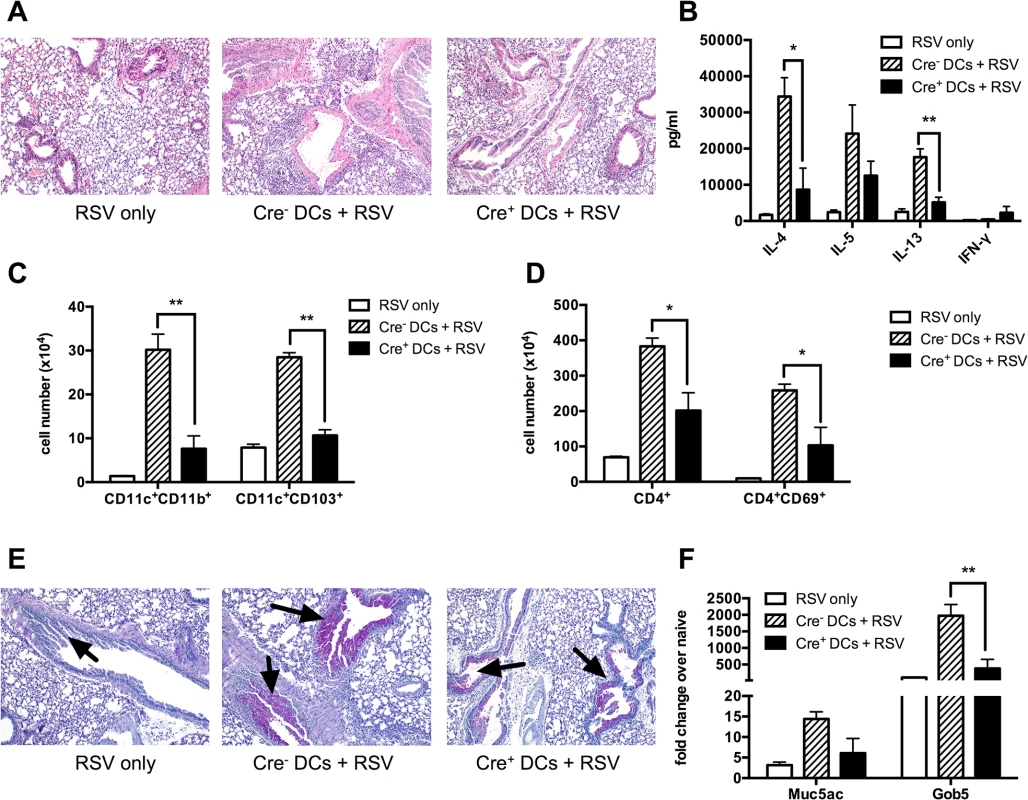
BMDCs from Kdm5bf/f-CD11c-Cre+ mice or Cre- controls were infected with RSV for 24 hours in vitro, then washed and administered intratracheally to WT C57BL/6 mice. After 7 days, mice were then infected with RSV, and samples were measured at 8 dpi. Mice infected with RSV alone were included as a control. (A) Lungs were removed and sections stained with H&E to visualize inflammation. (B) MLN were removed and restimulated in vitro for 48 hours with RSV. Supernatant proteins were measured by bioplex assay. (C) Lungs were homogenized into a single cell suspension and the number of CD11c+CD11b+ and CD11c+CD103+ DCs was quantified by flow cytometry. (D) CD4+ T cells and activated CD4+CD69+ T cells were measured by flow cytometry. (E) Lung sections were stained with PAS to visualize mucus production. Arrows indicate mucus production. (F) RNA was extracted from homogenized lung tissue and Muc5ac and Gob5 were measured by qPCR. *p<0.05, **p<0.01. Discussion
The objective of this study was to explore whether epigenetics are involved in determining the immune response during RSV infection. Previous studies have identified that DCs are a primary innate cell altered during RSV-induced pathogenesis, and that these cells play a central role in determining the nature of the immune responses. The concept that our immune system responses are influenced by environmental factors including microorganisms, pollution, diet and pathogen exposure is important as a backdrop for understanding disease progression [33–35]. Thus, we hypothesized that RSV infection of DCs would lead to the differential expression of epigenetic enzymes in these cells. The studies found that KDM5B, an H3K4 demethylase, was upregulated in DCs following RSV infection, but not when the cells were treated with ligands for TLR3 or TLR7, which are known to be important in recognition of the virus [22,23,36]. As H3K4me3 is an activation mark, it was further hypothesized that inhibiting KDM5B, which would remove methyl groups and thus remove the activation mark, would alter the activation state of the DC. Indeed, proinflammatory cytokines expressed by DCs were increased when KDM5B was blocked, which in vivo led to a suppression of the Th2 phenotype often associated with RSV infection. Thus, the present study identified several important concepts; 1) Epigenetic enzymes are a part of the mechanism by which a pathogen can modify the immune response; 2) Specific epigenetic enzymes have distinct effects on the function of DCs; 3) By identifying and specifically blocking epigenetic enzyme function a profound effect can be observed on the pulmonary immune environment. These results are summarized in Fig 7.
Fig. 7. Blocking KDM5B demethylation leads to increased cytokine production and repression of Th2 responses. 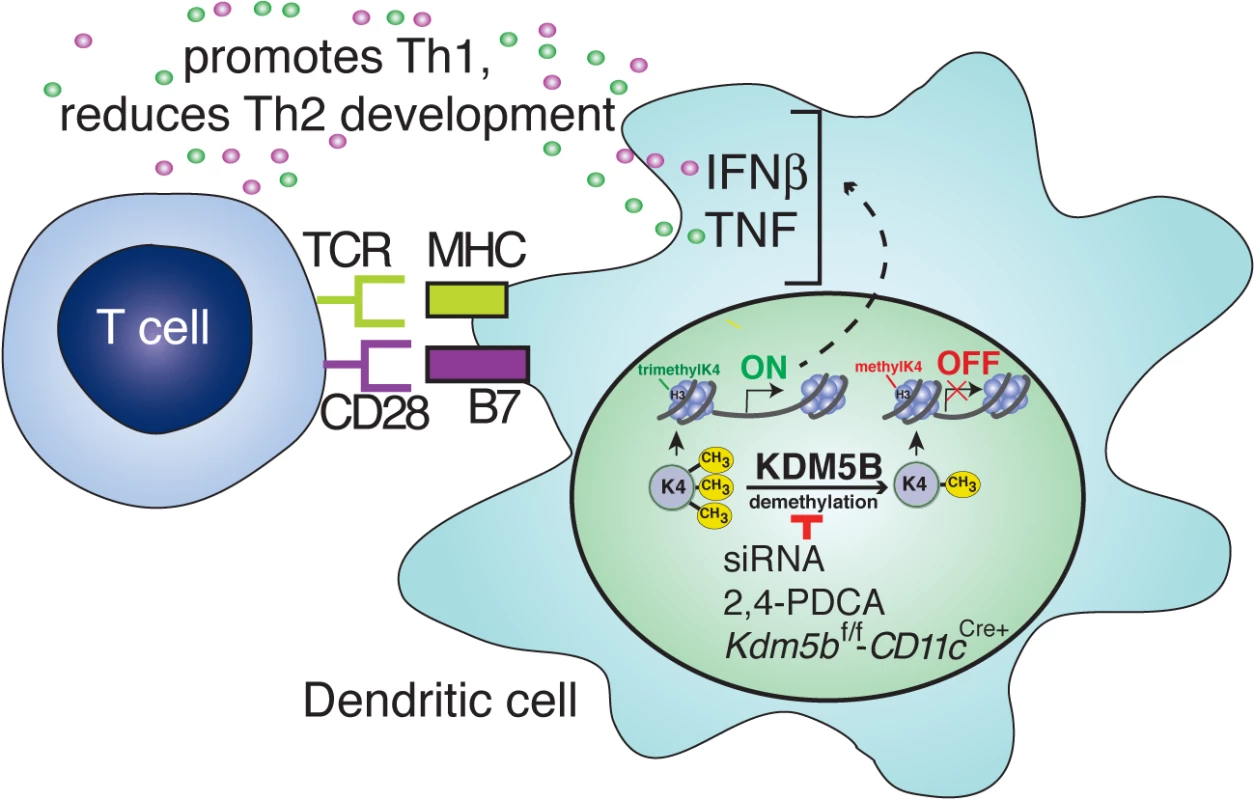
Following RSV infection, KDM5B is upregulated through unknown mechanisms. KDM5B demethylates H3K4 at the promoter regions of proinflammatory cytokines, leading to decreased transcription. Blocking Kdm5b by genetic means or siRNA silencing, or blocking KDM5B protein activity by chemical inhibition, leads to loss of demethylase activity, resulting in enhanced cytokine production from the DCs. Independent of MHC-II and costimulatory molecule surface expression on DCs, this increase in proinflammatory cytokines promotes Th1 responses and represses Th2 responses from the communicating T cells. Changes in the lung environment due to pathogen exposure have been well documented. Viral infections often induce long-term alterations in both immune function and lung physiology. For example, infection with influenza leads to a subsequent increase in susceptibility to bacterial infections, which is mediated in part by excessive IL-10 production in the lungs [37] and increased neutrophil apoptosis [38]. Conversely, influenza infection has been shown to be protective of the Th2 response associated with subsequent RSV infection in a mouse model [39]. Similarly, RSV infection also induces long-term changes in the lung environment, particularly when severe infection occurs early in life. Numerous studies have linked infection in infancy with childhood wheezing and the development of asthma [5,6]. While the mechanisms of this connection remain unclear, the cytokine environment that develops during infection may contribute to the pathology. In severe RSV infections during early childhood, the immune response in children requiring hospitalization is often characterized by the presence of Th2 cytokines and associated with increased eosinophil infiltration and mucus overproduction [40,41]. During early life, the immune system is predominantly biased toward a Th2 phenotype, and a Th1 balance does not develop until about one year of age [42], which may contribute to the altered immune response to RSV infection in the lungs of infants. The data presented in these studies highlight a potential Th2 reinforcing response that is induced by RSV in DC that would limit the ability of an individual to develop a more appropriate, less pathogenic anti-viral response. This specific epigenetic mechanism, elicited by KDM5B, may be but one contributing factor that influences RSV pathogenesis.
RSV is relatively poor at inducing type I IFNs, especially IFN-β, compared to other viruses including influenza [9,43–47]. The production of IFN-β from DCs has been shown to be important in directing DC maturation and cytokine production, indicating that increasing IFN-β production by DCs could alter the immune environment following RSV infection [48]. Previous studies have highlighted the importance of IFN-β production in developing an antigen-specific response to RSV, and have found that STAT1-deficient mice, which cannot initiate signaling from either type I or type II IFN, have significantly increased Th2 responses, as well as increased illness [26,49]. On the other hand, IFN-γ-deficient mice are protected, indicating that type I IFNs are important for developing an appropriate anti-viral response to RSV [26]. Furthermore, IFN-β and TNF-α are important in establishing a Th1 response in humans, thus increasing levels of these cytokines could help skew an effective immune response to RSV [50,51]. In support of these previous findings, the present study demonstrates that a primary consequence of removing KDMB from DC is the increase in both IFN-β and TNF-α, resulting in reduced Th2 pathology. Alternatively, increasing proinflammatory cytokines from DCs that lack functional KDM5B protein resulted in increased viral clearance from the lungs following primary RSV infection. The resulting decreased antigen levels in the lungs may potentially lead to DCs being able to preferentially drive a Th1 phenotype over Th2. However, as there were no differences in the antigen presenting ability of DCs deficient in Kdm5b, it is likely that the cytokines produced from the DCs are the primary factor that leads to decreased Th2 pathology.
Dendritic cells are the primary cell type responsible for stimulating CD4+ T cells and directing the adaptive immune response. Many of the long-term immunologic changes that occur in the lungs following RSV infection are due to alterations in DC function [27,28,46]. Although RSV nonstructural proteins (NS1 and NS2) have been specifically implicated in the inhibition of type I IFN production [9,52–57], the molecular mechanisms have not been fully explored. Recent studies in epigenetic mechanisms have led to a greater understanding of the molecular control of immune cells in the context of infectious agents. It has been shown that long-term changes in DC function and cytokine production following an inflammatory insult are due in part to epigenetic changes in the Il12 gene [17]. Others have demonstrated that the ability of DCs to prime Th1 responses is due to histone deacetylase activity [58]. More specifically, epigenetic enzymes can regulate the antiviral response by controlling the interferon pathway, especially IFN-stimulated genes [59]. This modulated immune effect was shown to be mediated by the G9a/GLP enzymatic complex [59]. In our studies, targeted gene arrays identified epigenetic enzymes that may contribute to the altered DC response following RSV infection. By using this unbiased approach, we targeted KDM5B as an important modulator of DC innate cytokine responses, as increased Kdm5b expression correlated with decreased Ifnb expression. By specifically inhibiting KDM5B and by generating a novel CD11c (DC) targeted KO mouse, studies identified that KDM5B has a critical role for altering DC function. Even more striking was the fact that the single enzyme alteration had a pathogenic impact on the anti-RSV response, including modifying T cell cytokine profiles and development of goblet cell metaplasia and mucus hypersecretion.
In summary, our studies identify a unique mechanism by which RSV infection influences how DCs are activated and subsequently activate the T cell responses leading to increased pulmonary pathogenesis. These studies used three methods to inhibit the expression or function of Kdm5b-siRNA knockdown, chemical inhibition and genetic deletion–and in all cases increased levels of innate cytokine production in DCs that subsequently led to a decrease in the Th2 response was observed. While previous attempts to create vaccines have led to increased Th2 pathology, these studies argue that future vaccine designs may need to consider the epigenetic programs that control pro-inflammatory cytokine production by DCs related to a protective immune response.
Materials and Methods
Ethics statement
All experiments on animal were performed in compliance with the guidelines of the Office of Laboratory Animal Welfare from the National Institutes of Health. Procedures were approved by the University Committee on the Use and Care of Animals at the University of Michigan (protocol PRO00004817, exp. 04/03/2016). All protocols using human cells were approved by the University of Michigan Institutional Review Board. For all experiments using human subjects, written informed consent was obtained from all donors.
Mice and infections
C57BL/6 mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA). Kdm5bfl/fl mice were previously described [31,60], and were crossed with B6.Cg-Tg(Itgax-cre)1-1Reiz/J (CD11c-Cre) mice (Jackson Laboratory). Mice were infected intratracheally with 1 x 105 pfu of RSV strain A2001/2-20, a clinical isolate originally from Vanderbilt University, was propagated as described [32]. Viral stocks were grown in Hep-2 cells and concentrations determined by plaque assay. In some experiments, cultured dendritic cells were infected with RSV for 24 hours, then thoroughly washed and 2.5 x 105 cells were transferred intratracheally into naïve mice.
Plaque assay
Whole lungs were harvested at two and four days post infection and were ground with a mortar and pestle. Supernatants were diluted and incubated with Vero cells for four days. RSV plaques were detected using a specific polyclonal antibody (Millipore, Billerica, MA).
Bone marrow-derived dendritic cell culture
Bone marrow was collected by flushing the femur and tibia of hind legs with PBS + 1% fetal calf serum (FCS). BMDCs were grown in RPMI 1640 supplemented with 10% FCS, L-glutamine, penicillin/streptomycin, non-essential amino acids, sodium pyruvate, 2-mercaptoethanol (ME) and 10 ng/ml of recombinant murine granulocyte macrophage-colony stimulating factor (GM-CSF; R&D Systems, Minneapolis, MN, USA). Cells were fed on days 3 and 5 with fresh GM-CSF. On day 6, cells were cultured with RSV (MOI = 1), 20 μg/ml of poly(I:C) or 1 μg/ml of imiquimod. For some experiments, cells were first treated for 24 hours with 1 mM 2,4-PDCA (Sigma, St. Louis, MO, USA), a chemical inhibitor of KDM5B [24,25], or with DMSO (0.1%) as a control, then were infected with RSV.
Monocyte-derived dendritic cell culture and co-culture with CD4+ T cells
Fifty to 70 ml of blood was taken by venous puncture. Peripheral blood mononuclear cells were isolated from heparinized blood by Ficoll-Paque (GE Healthcare) purification. Briefly, blood was diluted 1 : 1 with sodium chloride and tubes were centrifuged at 400g for 30 minutes with the brake off. Cells were harvested from the interface of the Ficoll layer, and were washed and enumerated. Monocytes were isolated using anti-CD14 microbeads, according to the manufacturer’s instruction (Miltenyi Biotec, San Diego, CA, USA). Cells were cultured in RPMI 1640 with 10% human serum and L-glutamine, pen/strep, non-essential amino acids, sodium pyruvate, 2-ME, 40 ng/ml of recombinant human IL-4 and 40 ng/ml of recombinant human GM-CSF (both from R&D Systems). Cytokines were replenished on days 3 and 6, and cells were used on day 7. For some experiments, CD4+ T cells were isolated from the CD14- fraction using the T cell isolation II kit (Miltenyi Biotec). Cells were then cultured with MoDCs in the presence or absence or RSV. At 48 hours, RNA was extracted and message levels of IFN-γ, IL-5 and IL-13 were determined by qPCR.
Transfection of dendritic cells with siRNA
BMDCs were transfected with siRNA at day 6 of culture. 1 μM of ON-TARGETplus or scrambled control siRNA (Dharmacon, Pittsburgh, PA, USA) was transfected using the Amaxa DC nucloefection kit and an Amaxa Nucleofector (Lonza Inc, Cologne, Germany). Cells were then cultured for 48 hours in complete medium and knock-down was assessed by qPCR. Cells were then infected with RSV for 24 hours.
RNA isolation and qPCR
RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) or RNeasy Mini Kit followed by the Cleanup Kit (Qiagen, Germantown, MD, USA) and following the manufacturers instructions. RNA isolated from tissues was first homogenized. Complementary DNA was synthesized using murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, CA, USA) and incubated at 37°C for one hour, followed by 95°C for 5 minutes to stop the reaction. Real-time quantitative PCR was multiplexed using Taqman primers with a FAM-conjugated probe and GAPDH with a VIC-conjugated probe (Applied Biosystems) to measure transcription of Il6, Tnf, Il13, Il4, Ifng and Kdm5b. Fold change was quantified using the 2-ΔΔCT method. Custom primers were designed to measure Ifnb, Muc5ac, Gob5 and RSV F, G and N RNA levels. All reactions were run on an ABI Prism 7500 Sequence Detection System or ViiA 7 Real Time PCR System (both from Applied Biosystems).
Histopathology
Lungs were removed at 8 days post infection. The large left lobe of each lung was inflated by injection with 4% formaldehyde. Lungs were embedded in paraffin, and 5 μm sections were sectioned and stained with hematoxylin and eosin (H&E) to visualize inflammatory cells or with periodic acid-Schiff stain (PAS) to visualize mucus production. To score mucus production, slides were evaluated by a blinded observer. Sections were scored based on the following scale: 1 –minimal, 2—slight, 3 –moderate, 4 –severe.
Flow cytometry
Lungs and mediastinal lymph nodes were removed and single cells were isolated by enzymatic digestion with 1 mg/ml collagenase A (Roche, Indianapolis, IN, USA) and 20 U/ml DNaseI (Sigma). Cells were resuspended in PBS with 1% FCS and Fc receptors were blocked with purified anti-CD16/32 (clone 93; BioLegend, San Diego, CA, USA). Surface markers were identified using antibodies (clones) against the following antigens, all from BioLegend, unless otherwise specified: CD11c (N418), CD11b (M1/70), CD103 (2E7), CD3 (145-2C11), CD4 (RM4-5), CD69 (H1.2F3), I-A/I-E (M5/114.15.2), CD80 (16-10A1), CD86 (GL-1), Gr-1 (RB6-8C5; eBiosciences, San Diego, CA, USA) and SiglecF (E50-2440; BD Biosciences, San Jose, CA, USA).
Cytokine production from lymph nodes
Mediatstinal lymph nodes were removed and single cells were isolated by enzymatic digestion. 5 x 105 cells were plated in 200 μl of complete medium and were restimulated with RSV for 48 hours. Supernatants were collected and levels of the cytokines IL-4, IL-5, IL-13 and IFN-γ were measured by Bioplex assay (Bio-Rad).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using an assay kit (Millipore) with minor modifications. Briefly, cells were fixed in 1% formaldehyde, then lysed in SDS buffer. Cells were then sonicated using a Branson Digital Sonifier 450 (VWR, West Chester, PA, USA) to create 200–1000 bp fragments. The lysate was clarified by centrifugation, and 5% of the supernatant was saved to measure the input DNA. The remaining chromatin was incubated with 1 μg of anti-H3K4me3 antibody (Abcam) or control IgG (Millipore) and incubated at 4°C with rotation overnight. Immune complexes were precipitated with salmon sperm DNA/protein A agarose beads. Crosslinking was reversed by incubation at 65°C and samples were treated with proteinase K. DNA was purified by phenol:chloroform:isoamyl alcohol separation and ethanol precipitation. Primers for the promoter regions of IFN-β, TNF-α and IL-6 were designed using Lasergene software and DNA was amplified by qPCR using SYBR Green buffer (Applied Biosystems).
Statistical analysis
Results are expressed and mean ± SE. Statistical significance was measured first by one-way or two-way ANOVA as appropriate, followed by a Student Neuman Keuhl’s post-hoc t test. A p value of <0.05 was considered significant.
Supporting Information
Zdroje
1. Shay DK, Holman RC, Newman RD, Liu LL, Stout JW, et al. Bronchiolitis-associated hospitalizations among US children, 1980–1996. JAMA 1999;282 : 1440–1446. 10535434
2. Simoes EA. Immunoprophylaxis of respiratory syncytial virus: global experience. Respir Res 2002;3 Suppl 1: S26–33. 12119055
3. Shay DK, Holman RC, Roosevelt GE, Clarke MJ, Anderson LJ. Bronchiolitis-associated mortality and estimates of respiratory syncytial virus-associated deaths among US children, 1979–1997. J Infect Dis 2001;183 : 16–22. 11076709
4. Howard TS, Hoffman LH, Stang PE, Simoes EA. Respiratory syncytial virus pneumonia in the hospital setting: length of stay, charges, and mortality. J Pediatr 2000;137 : 227–232. 10931416
5. Sigurs N, Bjarnason R, Sigurbergsson F, Kjellman B. Respiratory syncytial virus bronchiolitis in infancy is an important risk factor for asthma and allergy at age 7. Am J Respir Crit Care Med 2000;161 : 1501–1507. 10806145
6. Sigurs N, Gustafsson PM, Bjarnason R, Lundberg F, Schmidt S, et al. Severe respiratory syncytial virus bronchiolitis in infancy and asthma and allergy at age 13. Am J Respir Crit Care Med 2005;171 : 137–141. 15516534
7. Hall CB, Walsh EE, Long CE, Schnabel KC. Immunity to and frequency of reinfection with respiratory syncytial virus. J Infect Dis 1991;163 : 693–698. 2010624
8. Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, et al. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol 1969;89 : 422–434. 4305198
9. Ling Z, Tran KC, Teng MN. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J Virol 2009;83 : 3734–3742. doi: 10.1128/JVI.02434-08 19193793
10. Spann KM, Tran KC, Collins PL. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines. J Virol 2005;79 : 5353–5362. 15827150
11. Rudd BD, Smit JJ, Flavell RA, Alexopoulou L, Schaller MA, et al. Deletion of TLR3 alters the pulmonary immune environment and mucus production during respiratory syncytial virus infection. J Immunol 2006;176 : 1937–1942. 16424225
12. Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res 2007;17 : 691–707. 17567990
13. Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J 2011;30 : 4198–4210. doi: 10.1038/emboj.2011.295 21847099
14. Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, et al. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell 2014;55 : 347–360. doi: 10.1016/j.molcel.2014.06.005 24999238
15. Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity 2006;24 : 369–379. 16618596
16. Wei G, Wei L, Zhu J, Zang C, Hu-Li J, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 2009;30 : 155–167. doi: 10.1016/j.immuni.2008.12.009 19144320
17. Wen H, Dou Y, Hogaboam CM, Kunkel SL. Epigenetic regulation of dendritic cell-derived interleukin-12 facilitates immunosuppression after a severe innate immune response. Blood 2008;111 : 1797–1804. 18055863
18. Roesch A, Mueller AM, Stempfl T, Moehle C, Landthaler M, et al. RBP2-H1/JARID1B is a transcriptional regulator with a tumor suppressive potential in melanoma cells. Int J Cancer 2008;122 : 1047–1057. 17973255
19. Catteau A, Rosewell I, Solomon E, Taylor-Papadimitriou J. A short region of the promoter of the breast cancer associated PLU-1 gene can regulate transcription in vitro and in vivo. Int J Oncol 2004;25 : 5–16. 15201984
20. Madsen B, Tarsounas M, Burchell JM, Hall D, Poulsom R, et al. PLU-1, a transcriptional repressor and putative testis-cancer antigen, has a specific expression and localisation pattern during meiosis. Chromosoma 2003;112 : 124–132. 14579128
21. Shin HM, Kapoor VN, Guan T, Kaech SM, Welsh RM, et al. Epigenetic modifications induced by Blimp-1 Regulate CD8(+) T cell memory progression during acute virus infection. Immunity 2013;39 : 661–675. doi: 10.1016/j.immuni.2013.08.032 24120360
22. Lukacs NW, Smit JJ, Mukherjee S, Morris SB, Nunez G, et al. Respiratory virus-induced TLR7 activation controls IL-17-associated increased mucus via IL-23 regulation. J Immunol 2010;185 : 2231–2239. doi: 10.4049/jimmunol.1000733 20624950
23. Rudd BD, Burstein E, Duckett CS, Li X, Lukacs NW. Differential role for TLR3 in respiratory syncytial virus-induced chemokine expression. J Virol 2005;79 : 3350–3357. 15731229
24. Kristensen LH, Nielsen AL, Helgstrand C, Lees M, Cloos P, et al. Studies of H3K4me3 demethylation by KDM5B/Jarid1B/PLU1 reveals strong substrate recognition in vitro and identifies 2,4-pyridine-dicarboxylic acid as an in vitro and in cell inhibitor. FEBS J 2012;279 : 1905–1914. doi: 10.1111/j.1742-4658.2012.08567.x 22420752
25. Sayegh J, Cao J, Zou MR, Morales A, Blair LP, et al. Identification of small molecule inhibitors of Jumonji AT-rich interactive domain 1B (JARID1B) histone demethylase by a sensitive high throughput screen. J Biol Chem 2013;288 : 9408–9417. doi: 10.1074/jbc.M112.419861 23408432
26. Durbin JE, Johnson TR, Durbin RK, Mertz SE, Morotti RA, et al. The role of IFN in respiratory syncytial virus pathogenesis. J Immunol 2002;168 : 2944–2952. 11884466
27. Reed M, Morris SH, Jang S, Mukherjee S, Yue Z, et al. Autophagy-inducing protein beclin-1 in dendritic cells regulates CD4 T cell responses and disease severity during respiratory syncytial virus infection. J Immunol 2013;191 : 2526–2537. doi: 10.4049/jimmunol.1300477 23894198
28. Jang S, Smit J, Kallal LE, Lukacs NW. Respiratory syncytial virus infection modifies and accelerates pulmonary disease via DC activation and migration. J Leukoc Biol 2013;94 : 5–15. doi: 10.1189/jlb.0412195 23293372
29. GeurtsvanKessel CH, Lambrecht BN. Division of labor between dendritic cell subsets of the lung. Mucosal Immunol 2008;1 : 442–450. doi: 10.1038/mi.2008.39 19079211
30. Sung SS, Fu SM, Rose CE Jr., Gaskin F, Ju ST, et al. A major lung CD103 (alphaE)-beta7 integrin-positive epithelial dendritic cell population expressing Langerin and tight junction proteins. J Immunol 2006;176 : 2161–2172. 16455972
31. Albert M, Schmitz SU, Kooistra SM, Malatesta M, Morales Torres C, et al. The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet 2013;9: e1003461. doi: 10.1371/journal.pgen.1003461 23637629
32. Stokes KL, Chi MH, Sakamoto K, Newcomb DC, Currier MG, et al. Differential pathogenesis of respiratory syncytial virus clinical isolates in BALB/c mice. J Virol 2011;85 : 5782–5793. doi: 10.1128/JVI.01693-10 21471228
33. Suarez-Alvarez B, Rodriguez RM, Fraga MF, Lopez-Larrea C. DNA methylation: a promising landscape for immune system-related diseases. Trends Genet 2012;28 : 506–514. doi: 10.1016/j.tig.2012.06.005 22824525
34. Rodriguez-Cortez VC, Hernando H, de la Rica L, Vento R, Ballestar E. Epigenomic deregulation in the immune system. Epigenomics 2011;3 : 697–713. doi: 10.2217/epi.11.99 22126290
35. Adhya D, Basu A. Epigenetic modulation of host: new insights into immune evasion by viruses. J Biosci 2010;35 : 647–663. 21289446
36. Groskreutz DJ, Monick MM, Powers LS, Yarovinsky TO, Look DC, et al. Respiratory syncytial virus induces TLR3 protein and protein kinase R, leading to increased double-stranded RNA responsiveness in airway epithelial cells. J Immunol 2006;176 : 1733–1740. 16424203
37. van der Sluijs KF, van Elden LJ, Nijhuis M, Schuurman R, Pater JM, et al. IL-10 is an important mediator of the enhanced susceptibility to pneumococcal pneumonia after influenza infection. J Immunol 2004;172 : 7603–7609. 15187140
38. Colamussi ML, White MR, Crouch E, Hartshorn KL. Influenza A virus accelerates neutrophil apoptosis and markedly potentiates apoptotic effects of bacteria. Blood 1999;93 : 2395–2403. 10090951
39. Walzl G, Tafuro S, Moss P, Openshaw PJ, Hussell T. Influenza virus lung infection protects from respiratory syncytial virus-induced immunopathology. J Exp Med 2000;192 : 1317–1326. 11067880
40. Kristjansson S, Bjarnarson SP, Wennergren G, Palsdottir AH, Arnadottir T, et al. Respiratory syncytial virus and other respiratory viruses during the first 3 months of life promote a local TH2-like response. J Allergy Clin Immunol 2005;116 : 805–811. 16210054
41. Legg JP, Hussain IR, Warner JA, Johnston SL, Warner JO. Type 1 and type 2 cytokine imbalance in acute respiratory syncytial virus bronchiolitis. Am J Respir Crit Care Med 2003;168 : 633–639. 12773328
42. Siegrist CA. Neonatal and early life vaccinology. Vaccine 2001;19 : 3331–3346. 11348697
43. Jie Z, Dinwiddie DL, Senft AP, Harrod KS. Regulation of STAT signaling in mouse bone marrow derived dendritic cells by respiratory syncytial virus. Virus Res 2011;156 : 127–133. doi: 10.1016/j.virusres.2011.01.007 21255624
44. Shingai M, Azuma M, Ebihara T, Sasai M, Funami K, et al. Soluble G protein of respiratory syncytial virus inhibits Toll-like receptor 3/4-mediated IFN-beta induction. Int Immunol 2008;20 : 1169–1180. doi: 10.1093/intimm/dxn074 18611945
45. Jewell NA, Vaghefi N, Mertz SE, Akter P, Peebles RS Jr., et al. Differential type I interferon induction by respiratory syncytial virus and influenza a virus in vivo. J Virol 2007;81 : 9790–9800. 17626092
46. Guerrero-Plata A, Kolli D, Hong C, Casola A, Garofalo RP. Subversion of pulmonary dendritic cell function by paramyxovirus infections. J Immunol 2009;182 : 3072–3083. doi: 10.4049/jimmunol.0802262 19234204
47. Ramaswamy M, Shi L, Monick MM, Hunninghake GW, Look DC. Specific inhibition of type I interferon signal transduction by respiratory syncytial virus. Am J Respir Cell Mol Biol 2004;30 : 893–900. 14722224
48. Rudd BD, Luker GD, Luker KE, Peebles RS, Lukacs NW. Type I interferon regulates respiratory virus infected dendritic cell maturation and cytokine production. Viral Immunol 2007;20 : 531–540. 18158727
49. Hashimoto K, Durbin JE, Zhou W, Collins RD, Ho SB, et al. Respiratory syncytial virus infection in the absence of STAT 1 results in airway dysfunction, airway mucus, and augmented IL-17 levels. J Allergy Clin Immunol 2005;116 : 550–557. 16159623
50. Nagai T, Devergne O, Mueller TF, Perkins DL, van Seventer JM, et al. Timing of IFN-beta exposure during human dendritic cell maturation and naive Th cell stimulation has contrasting effects on Th1 subset generation: a role for IFN-beta-mediated regulation of IL-12 family cytokines and IL-18 in naive Th cell differentiation. J Immunol 2003;171 : 5233–5243. 14607924
51. Henig N, Avidan N, Mandel I, Staun-Ram E, Ginzburg E, et al. Interferon-beta induces distinct gene expression response patterns in human monocytes versus T cells. PLoS One 2013;8: e62366. doi: 10.1371/journal.pone.0062366 23626809
52. Boyapalle S, Wong T, Garay J, Teng M, San Juan-Vergara H, et al. Respiratory syncytial virus NS1 protein colocalizes with mitochondrial antiviral signaling protein MAVS following infection. PLoS One 2012;7: e29386. doi: 10.1371/journal.pone.0029386 22383950
53. Munir S, Hillyer P, Le Nouen C, Buchholz UJ, Rabin RL, et al. Respiratory syncytial virus interferon antagonist NS1 protein suppresses and skews the human T lymphocyte response. PLoS Pathog 2011;7: e1001336. doi: 10.1371/journal.ppat.1001336 21533073
54. Munir S, Le Nouen C, Luongo C, Buchholz UJ, Collins PL, et al. Nonstructural proteins 1 and 2 of respiratory syncytial virus suppress maturation of human dendritic cells. J Virol 2008;82 : 8780–8796. doi: 10.1128/JVI.00630-08 18562519
55. Moore EC, Barber J, Tripp RA. Respiratory syncytial virus (RSV) attachment and nonstructural proteins modify the type I interferon response associated with suppressor of cytokine signaling (SOCS) proteins and IFN-stimulated gene-15 (ISG15). Virol J 2008;5 : 116. doi: 10.1186/1743-422X-5-116 18851747
56. Spann KM, Tran KC, Chi B, Rabin RL, Collins PL. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages [corrected]. J Virol 2004;78 : 4363–4369. 15047850
57. Teng MN, Whitehead SS, Bermingham A, St Claire M, Elkins WR, et al. Recombinant respiratory syncytial virus that does not express the NS1 or M2-2 protein is highly attenuated and immunogenic in chimpanzees. J Virol 2000;74 : 9317–9321. 10982380
58. Brogdon JL, Xu Y, Szabo SJ, An S, Buxton F, et al. Histone deacetylase activities are required for innate immune cell control of Th1 but not Th2 effector cell function. Blood 2007;109 : 1123–1130. 17008546
59. Fang TC, Schaefer U, Mecklenbrauker I, Stienen A, Dewell S, et al. Histone H3 lysine 9 di-methylation as an epigenetic signature of the interferon response. J Exp Med 2012;209 : 661–669. doi: 10.1084/jem.20112343 22412156
60. Schmitz SU, Albert M, Malatesta M, Morey L, Johansen JV, et al. Jarid1b targets genes regulating development and is involved in neural differentiation. Embo J 2011;30 : 4586–4600. doi: 10.1038/emboj.2011.383 22020125
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage TraffickingČlánek An Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived NeuronsČlánek Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPsČlánek Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African TrypanosomiasisČlánek Peptidoglycan Branched Stem Peptides Contribute to Virulence by Inhibiting Pneumolysin ReleaseČlánek HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T CellsČlánek Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 6- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Introducing “Research Matters”
- Exploring Host–Pathogen Interactions through Biological Control
- Analysis of Bottlenecks in Experimental Models of Infection
- Expected and Unexpected Features of the Newly Discovered Bat Influenza A-like Viruses
- Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage Trafficking
- Recombinant Murine Gamma Herpesvirus 68 Carrying KSHV G Protein-Coupled Receptor Induces Angiogenic Lesions in Mice
- TRIM30α Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING
- Targeting Human Transmission Biology for Malaria Elimination
- Two Cdc2 Kinase Genes with Distinct Functions in Vegetative and Infectious Hyphae in
- An Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived Neurons
- Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPs
- Virulence Factors of Induce Both the Unfolded Protein and Integrated Stress Responses in Airway Epithelial Cells
- Peptide-MHC-I from Endogenous Antigen Outnumber Those from Exogenous Antigen, Irrespective of APC Phenotype or Activation
- Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African Trypanosomiasis
- An Ultrasensitive Mechanism Regulates Influenza Virus-Induced Inflammation
- The Role of Human Transportation Networks in Mediating the Genetic Structure of Seasonal Influenza in the United States
- Host Delivery of Favorite Meals for Intracellular Pathogens
- Complement-Opsonized HIV-1 Overcomes Restriction in Dendritic Cells
- Inter-Seasonal Influenza is Characterized by Extended Virus Transmission and Persistence
- A Critical Role for CLSP2 in the Modulation of Antifungal Immune Response in Mosquitoes
- Twilight, a Novel Circadian-Regulated Gene, Integrates Phototropism with Nutrient and Redox Homeostasis during Fungal Development
- Surface-Associated Lipoproteins Link Virulence to Colitogenic Activity in IL-10-Deficient Mice Independent of Their Expression Levels
- Latent Membrane Protein LMP2A Impairs Recognition of EBV-Infected Cells by CD8+ T Cells
- Bank Vole Prion Protein As an Apparently Universal Substrate for RT-QuIC-Based Detection and Discrimination of Prion Strains
- Neuronal Subtype and Satellite Cell Tropism Are Determinants of Varicella-Zoster Virus Virulence in Human Dorsal Root Ganglia Xenografts
- Molecular Basis for the Selective Inhibition of Respiratory Syncytial Virus RNA Polymerase by 2'-Fluoro-4'-Chloromethyl-Cytidine Triphosphate
- Structure of the Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome
- Activated Brain Endothelial Cells Cross-Present Malaria Antigen
- Fungal Morphology, Iron Homeostasis, and Lipid Metabolism Regulated by a GATA Transcription Factor in
- Peptidoglycan Branched Stem Peptides Contribute to Virulence by Inhibiting Pneumolysin Release
- A Macrophage Subversion Factor Is Shared by Intracellular and Extracellular Pathogens
- A Novel AT-Rich DNA Recognition Mechanism for Bacterial Xenogeneic Silencer MvaT
- Reovirus FAST Proteins Drive Pore Formation and Syncytiogenesis Using a Novel Helix-Loop-Helix Fusion-Inducing Lipid Packing Sensor
- The Role of ExoS in Dissemination of during Pneumonia
- IRF-5-Mediated Inflammation Limits CD8 T Cell Expansion by Inducing HIF-1α and Impairing Dendritic Cell Functions during Infection
- Discordant Impact of HLA on Viral Replicative Capacity and Disease Progression in Pediatric and Adult HIV Infection
- Crystal Structure of USP7 Ubiquitin-like Domains with an ICP0 Peptide Reveals a Novel Mechanism Used by Viral and Cellular Proteins to Target USP7
- HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells
- HPV16 Down-Regulates the Insulin-Like Growth Factor Binding Protein 2 to Promote Epithelial Invasion in Organotypic Cultures
- The νSaα Specific Lipoprotein Like Cluster () of . USA300 Contributes to Immune Stimulation and Invasion in Human Cells
- RSV-Induced H3K4 Demethylase KDM5B Leads to Regulation of Dendritic Cell-Derived Innate Cytokines and Exacerbates Pathogenesis
- Leukocidin A/B (LukAB) Kills Human Monocytes via Host NLRP3 and ASC when Extracellular, but Not Intracellular
- Border Patrol Gone Awry: Lung NKT Cell Activation by Exacerbates Tularemia-Like Disease
- The Curious Road from Basic Pathogen Research to Clinical Translation
- From Cell and Organismal Biology to Drugs
- Adenovirus Tales: From the Cell Surface to the Nuclear Pore Complex
- A 21st Century Perspective of Poliovirus Replication
- Is Development of a Vaccine against Feasible?
- Waterborne Viruses: A Barrier to Safe Drinking Water
- Battling Phages: How Bacteria Defend against Viral Attack
- Archaea in and on the Human Body: Health Implications and Future Directions
- Degradation of Human PDZ-Proteins by Human Alphapapillomaviruses Represents an Evolutionary Adaptation to a Novel Cellular Niche
- Natural Variants of the KPC-2 Carbapenemase have Evolved Increased Catalytic Efficiency for Ceftazidime Hydrolysis at the Cost of Enzyme Stability
- Potent Cell-Intrinsic Immune Responses in Dendritic Cells Facilitate HIV-1-Specific T Cell Immunity in HIV-1 Elite Controllers
- The Mammalian Cell Cycle Regulates Parvovirus Nuclear Capsid Assembly
- Host Reticulocytes Provide Metabolic Reservoirs That Can Be Exploited by Malaria Parasites
- The Proteome of the Isolated Containing Vacuole Reveals a Complex Trafficking Platform Enriched for Retromer Components
- NK-, NKT- and CD8-Derived IFNγ Drives Myeloid Cell Activation and Erythrophagocytosis, Resulting in Trypanosomosis-Associated Acute Anemia
- Successes and Challenges on the Road to Cure Hepatitis C
- BRCA1 Regulates IFI16 Mediated Nuclear Innate Sensing of Herpes Viral DNA and Subsequent Induction of the Innate Inflammasome and Interferon-β Responses
- A Structural and Functional Comparison Between Infectious and Non-Infectious Autocatalytic Recombinant PrP Conformers
- Phosphorylation of the Peptidoglycan Synthase PonA1 Governs the Rate of Polar Elongation in Mycobacteria
- Human Immunodeficiency Virus Type 1 Nef Inhibits Autophagy through Transcription Factor EB Sequestration
- Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes
- EBV BART MicroRNAs Target Multiple Pro-apoptotic Cellular Genes to Promote Epithelial Cell Survival
- Single-Cell and Single-Cycle Analysis of HIV-1 Replication
- TRIM32 Senses and Restricts Influenza A Virus by Ubiquitination of PB1 Polymerase
- The Herpes Simplex Virus Protein pUL31 Escorts Nucleocapsids to Sites of Nuclear Egress, a Process Coordinated by Its N-Terminal Domain
- Host Transcriptional Response to Influenza and Other Acute Respiratory Viral Infections – A Prospective Cohort Study
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells
- Battling Phages: How Bacteria Defend against Viral Attack
- A 21st Century Perspective of Poliovirus Replication
- Adenovirus Tales: From the Cell Surface to the Nuclear Pore Complex
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



