-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaEpigenetic Control of O-Antigen Chain Length: A Tradeoff between Virulence and Bacteriophage Resistance
A tradeoff can increase the adaptive capacity of an organism at the expense of lowering the fitness conferred by specific traits. This study describes a tradeoff that confers bacteriophage resistance in Salmonella enterica at the expense of reducing its pathogenic capacity. Phase variation of the opvAB operon creates two subpopulations of bacterial cells, each with a distinct lipopolysaccharide structure. One subpopulation is large and virulent but sensitive to phages that use the lipopolysaccharide O-antigen as receptor, while the other is small and avirulent but phage resistant. In the presence of a phage that targets the O-antigen, only the avirulent subpopulation survives. However, phase variation permits resuscitation of the virulent opvABOFF subpopulation as soon as phage challenge ceases. This transient tradeoff may illustrate the adaptive value of epigenetic mechanisms that generate bacterial subpopulations in a reversible manner.
Published in the journal: . PLoS Genet 11(11): e32767. doi:10.1371/journal.pgen.1005667
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005667Summary
A tradeoff can increase the adaptive capacity of an organism at the expense of lowering the fitness conferred by specific traits. This study describes a tradeoff that confers bacteriophage resistance in Salmonella enterica at the expense of reducing its pathogenic capacity. Phase variation of the opvAB operon creates two subpopulations of bacterial cells, each with a distinct lipopolysaccharide structure. One subpopulation is large and virulent but sensitive to phages that use the lipopolysaccharide O-antigen as receptor, while the other is small and avirulent but phage resistant. In the presence of a phage that targets the O-antigen, only the avirulent subpopulation survives. However, phase variation permits resuscitation of the virulent opvABOFF subpopulation as soon as phage challenge ceases. This transient tradeoff may illustrate the adaptive value of epigenetic mechanisms that generate bacterial subpopulations in a reversible manner.
Introduction
The study of differentiation in bacterial species that undergo developmental programs has played a historic role in biology [1,2,3]. In addition, phenotypic differences between colonies [4] and within colonies [5,6] were described many years ago in bacterial species that do not undergo development. Despite their technical limitations, these early studies contributed to bring about the idea that phenotypic heterogeneity might be a common phenomenon in the bacterial world [7]. This view has been confirmed by single cell analysis technologies [8,9,10,11,12]. Furthermore, theoretical analysis has provided evidence that phenotypic heterogeneity can have adaptive value, especially in hostile or changing environments [13,14,15]. In certain cases, the adaptive value of subpopulation formation is illustrated by experimental evidence [16,17,18].
Formation of bacterial lineages is governed by diverse mechanisms, including programmed genetic rearrangement [19] and contraction or expansion of DNA repeats at genome regions [20,21]. In other cases, however, lineage formation is controlled by epigenetic mechanisms: certain cell-to-cell differences serve as physiological signals, and signal propagation by a feedback loop generates an inheritable phenotype [12,22]. Cell-to-cell differences can be a consequence of environmental inputs or result from the noise intrinsical to many cellular processes [10,12,15]. In turn, the feedback loops that propagate the initial state beyond can be relatively simple or involve complex mechanisms like the formation of inheritable DNA adenine methylation patterns in the genome [12,23,24]. Some feedback loops are stable enough to cause bistability, the bifurcation of a bacterial population into two distinct phenotypic states [22].
If a feedback loop is metastable, reversion of the epigenetic state will occur after a certain number of cell divisions. Reversible bistability is usually known as phase variation, and typically involves reversible switching of gene expression from OFF to ON or from low to high expression [25,26,27]. Examples of phase variation have been described mostly in bacterial pathogens, and subpopulation formation is frequently viewed as a strategy that may facilitate evasion of the immune system during infection of animals [25,26]. This view is supported by the observation that phase-variable loci often encode envelope components or proteins involved in modification of the bacterial envelope [25,26].
Some phase-variable envelope modifications controlled by DNA adenine methylation play roles in bacteriophage resistance. For instance, phase variation in the gtrABC1 cluster protects S. enterica against the T5-like phage SPC35, probably by an indirect mechanism [28]. In Haemophilus influenzae, DNA adenine methylation controls phase-variable resistance to bacteriophage HP1c1 but the underlying mechanism remains hypothetical [29]. Phase variation can also contribute to phage resistance without alteration of the bacterial surface. For instance, certain genes encoding restriction-modification systems show phase variation [30,31].
In this study, we describe a phase variation system that confers resistance to bacteriophages that use the lipopolysaccharide (LPS) O-antigen as receptor. The genome of Salmonella enterica contains a horizontally-acquired locus, known as opvAB or STM2209-STM2208 [32]. The opvA and opvB genes form a bicistronic operon [32] and encode inner membrane proteins [32]. OpvA is a small peptide of 34 amino acids, and OpvB is a larger protein of 221 amino acids with homology to the Wzz superfamily of regulators of LPS O-antigen chain length [32]. We show that expression of the S. enterica opvAB operon confers resistance to bacteriophages P22 (Podoviridae), 9NA (Siphoviridae), and Det7 (Myoviridae) by modification of the phage receptor, the LPS O-antigen. Because expression of opvAB is phase-variable, bacteriophage resistance occurs in the subpopulation of opvABON cells only. This subpopulation, which is extremely small, preadapts Salmonella to survive phage challenge albeit at the cost of reduced virulence. However, because the opvABON state is reversible, the virulence payoff is temporary, and a virulent bacterial population resuscitates as soon as phage challenge ceases.
Results
Localization of the OpvA and OpvB proteins in the S. enterica envelope
OpvA and OpvB were previously shown to be inner membrane proteins involved in LPS synthesis [32]. Because the LPS is known to have a helical distribution in the cell envelope [33], the OpvA and OpvB subcellular localization was investigated. For this purpose, a chromosomal opvB::mCherry fusion was constructed downstream of the opvB gene (so that the strain remains OpvAB+). In a wild type background, expression of opvB::mCherry was low in most cells (Fig 1A). However, rare cells with high levels of expression of opvB::mCherry were detected (Fig 1A), an observation consistent with the occurrence of phase variation skewed towards the OFF state [32]. Expression of opvB::mCherry was also monitored in an opvAB-constitutive (opvABON) strain engineered by elimination of GATC sites upstream of the opvAB promoter [32]. In an opvABON background, all cells displayed high levels of fluorescence, similar to those of the rare fluorescent cells visualized in a wild type background (Fig 1B). In fluorescent cells, OpvB was seen forming helical intertwining ribbons in the inner membrane (Fig 1A and 1B).
Fig. 1. Analysis of OpvA and OpvB localization by fluorescence microscopy. 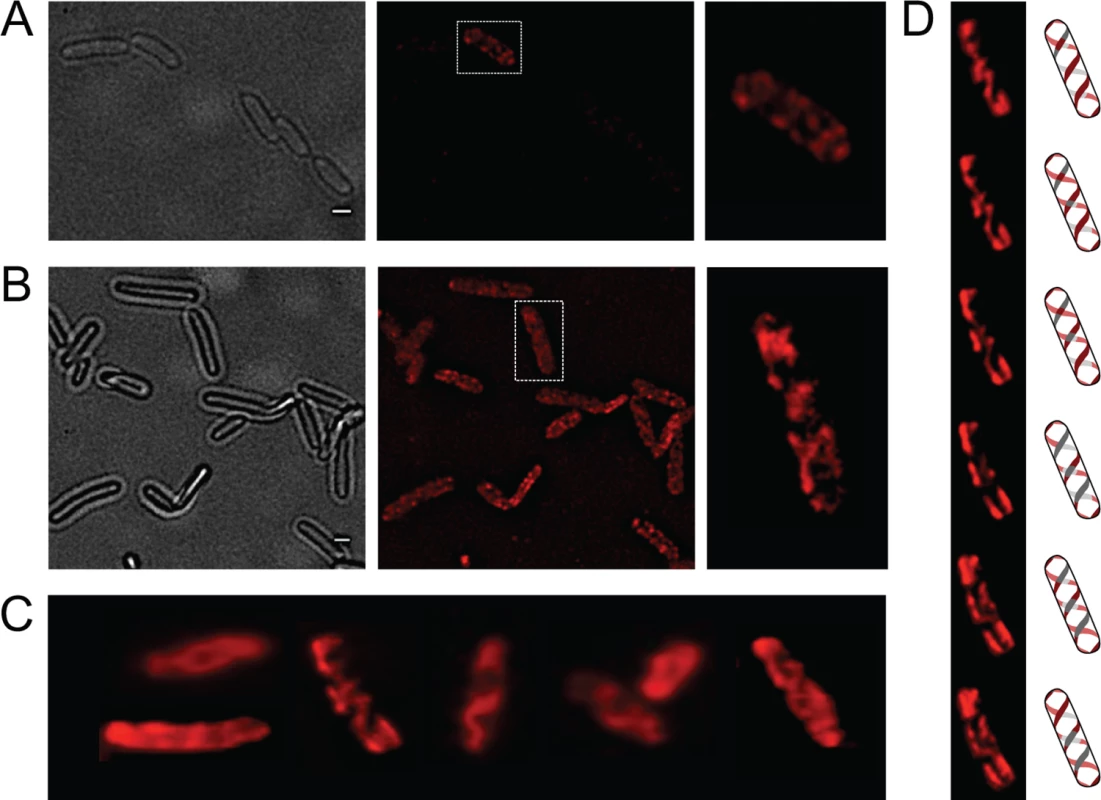
A. Localization of OpvB-mCherry in a wild type background. B. Localization of OpvB-mCherry in an opvABON strain. In both panels, cells enclosed in boxes are shown with higher magnification on the right. C. Localization of plasmid-borne OpvA-mCherry. D. Z-stacks of a single cell from panel C accompanied by an idealized representation of the ribbon-like protein distribution. Scale bar: 1 μm. The subcellular distribution of OpvA was examined using a plasmid-borne opvA::mCherry fusion. This experimental choice was based on the consideration that construction of an mCherry fusion in the upstream gene opvA would likely prevent opvB expression because of a polarity effect. In the strain carrying plasmid-borne opvA::mCherry, intense fluorescence was observed in all cells (Fig 1C), presumably because opvA::mCherry overexpression from the multicopy plasmid abolished phase variation. This construction was useful, however, to permit clear-cut observation of helical intertwining ribbons formed by OpvA (Fig 1D). The evidence that OpvA and OpvB may have a similar or identical distribution in the bacterial envelope is consistent with the physical interaction previously described between OpvA and OpvB [32].
Roles of OpvA and OpvB in control of O-antigen chain length
Constitutive expression of opvAB leads to the production of a particular form of O-antigen in the S. enterica LPS, with a modal length of 3–8 repeat units [32]. A diagram of LPS structure is presented in S1 Fig, together with an electrophoretic separation of O-antigen chains and a diagram of the differences in LPS structure between opvABOFF and opvABON pubpopulations. To investigate the role of individual OpvA and OpvB proteins in control of O-antigen chain length, non-polar mutations in opvA and opvB were constructed in the wild type and in an opvABON background. In the wild type, lack of either OpvA or OpvB did not alter the electrophoretic profile of LPS (Fig 2), an observation consistent with two known facts: the subpopulation of cells that express opvAB in wild type Salmonella is very small [32], and an OpvAB−mutant displays an LPS profile identical to that of the wild type [32]. In contrast, OpvA−opvBON and opvAON OpvB−mutants showed differences with the parental opvABON strain and also with the wild type:
Absence of OpvB (opvAON) yielded a seemingly disorganized LPS with no clear modal length (Fig 2), reminiscent of the LPS produced in the absence of the modal length regulators WzzST and WzzfepE [34,35,36].
Absence of OpvA (opvBON) yielded an LPS with the modal lengths typically conferred by WzzST and WzzfepE [34] but also showed a preferred opvABON-like modal length in the lower weight band region (Fig 2).
Fig. 2. Analysis of the roles of OpvA and OpvB in the control of O-antigen chain length. 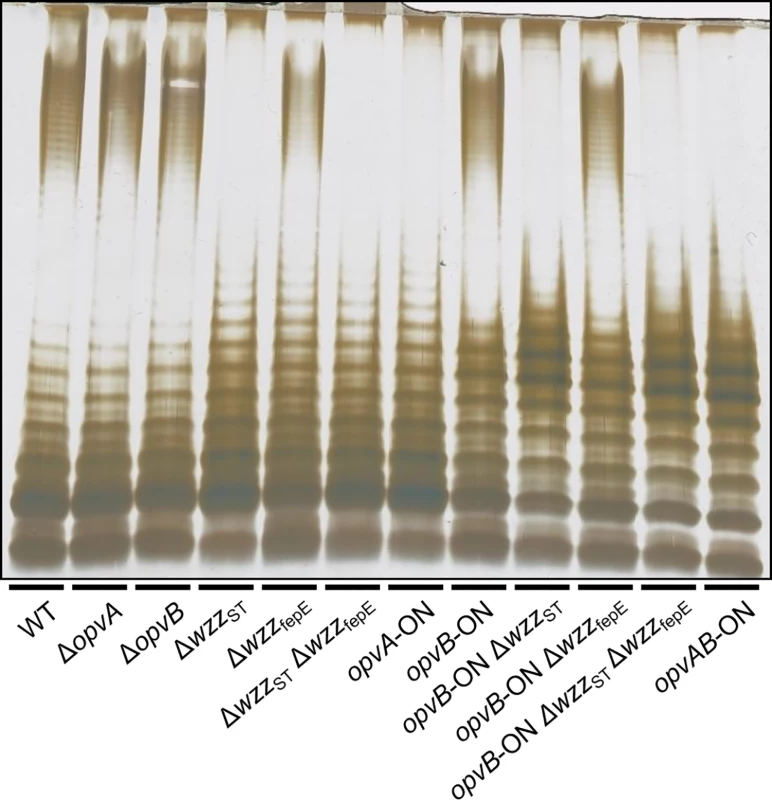
LPS profiles of S. enterica strains carrying mutations in opvAB and O-antigen length regulator genes wzzST and wzzfepE, as observed by electrophoresis and silver staining. These observations suggested that the function of OpvA might be to prevent the formation of normal O-antigen so that OpvB could then impose its preferred modal length. To test this hypothesis, LPS structure was analyzed in an OpvA−opvBON background in the absence of either WzzST or WzzfepE. The results support the view that OpvB needs OpvA to prevent O-antigen formation by customary modal length regulators. In the absence of WzzST, OpvB alone was able to produce an O-antigen similar to that found in the opvABON strain (Fig 2). In contrast, lack of WzzfepE did not seem to facilitate OpvB function, suggesting that OpvB may mainly compete with WzzST. This preference may be related to the fact that both WzzST and OpvB convey relatively short preferred modal lengths: 3–8 for OpvB [32] and 16–35 for WzzST [36,37,38] compared with >100 for WzzfepE [34].
Selection of opvABON S. enterica cells upon bacteriophage challenge
The LPS O-antigen is a typical receptor for bacteriophages [39] and modification of the O-antigen can confer bacteriophage resistance [40]. On these grounds, we tested whether opvAB expression increased Salmonella resistance to the virulent phages 9NA [41,42] and Det7 [43,44]. We also tested the historic phage P22, using a virulent mutant to avoid lysogeny [45]. Three strains (wild type, opvABON and ΔopvAB) were challenged with 9NA, Det7, and P22, which belong to different bacteriophage families and use the O-antigen as receptor. The experiments shown in Fig 3 were carried out by inoculating an exponential culture of S. enterica with an aliquot of a phage suspension at a multiplicity of infection (MOI) >10, and monitoring bacterial growth afterwards. The results can be summarized as follows:
Growth of the opvABON strain was not affected by the presence of 9NA, Det7, or P22, suggesting that the strain was resistant to these bacteriophages (S2 Fig).
A culture of the wild type strain became clear 1–2 hours after P22 infection, suggesting that cell lysis had occurred. However, bacterial growth was observed around 4 hours after infection, and was interpreted as occurrence of P22 resistance. Infection with either 9NA or Det7 did not cause clearing but growth retardation, and growth resumed 4 hours after infection (Fig 3).
Cultures of the ΔopvAB strain infected with 9NA, Det7, or P22 became clear or almost clear. Growth was detected later, albeit with significant delay compared with the wild type. Because the wild type, the opvABON strain and the ΔopvAB strain show similar or identical growth rates in LB broth (S2 Fig), the explanation of this phenomenon is that growth of the ΔopvAB strain in the presence of 9NA, Det7, or P22 selects phage-resistant mutants (see below).
Fig. 3. Effect of phage challenge on S. enterica growth and LPS structure. 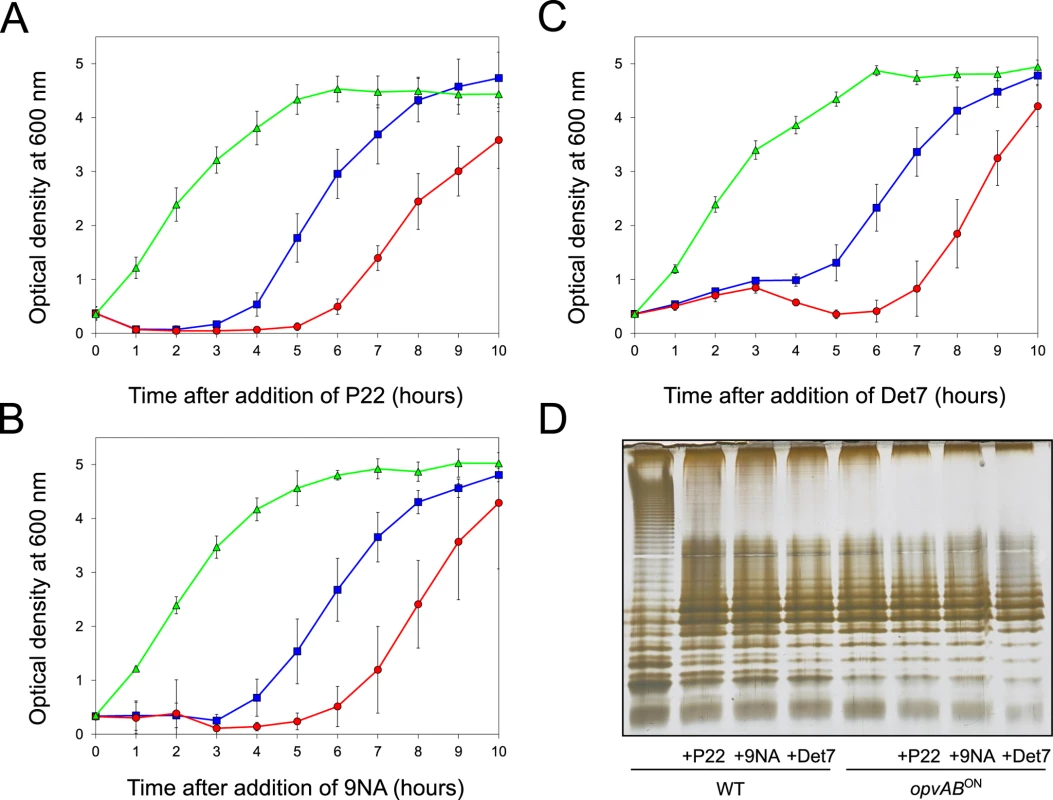
Growth of the wild type strain (blue squares), an opvABON strain (green triangles) and a ΔopvAB strain (red circles) in LB + P22 (A), LB + 9NA (B), and LB + Det7 (C). Values are averages and standard deviations from ≥ 6 independent experiments. D. LPS structure of the wild type and opvABON strains after growth in LB, LB + P22, LB + 9NA, and LB + Det7. A tentative interpretation of these observations was that the wild type strain contained a subpopulation of opvABON cells that survived phage challenge. Because opvAB phase variation is skewed towards the OFF state [32], the small size of the opvABON subpopulation and the regular formation of phage-sensitive opvABOFF cells caused growth retardation (albeit to different degrees depending on the phage). In contrast, the opvABON strain grew normally, an observation consistent with the occurrence of phage resistance in the entire bacterial population. This interpretation was supported by analysis of the LPS profiles of wild type and opvABON strains grown in the presence of P22, 9NA, and Det7 until stationary phase (OD600 ~4) (Fig 3). After phage challenge, the wild type contained an LPS different from the LPS found in LB (Fig 3D), and similar or identical to the LPS found in the opvABON strain (Fig 2; see also [32]). In contrast, the LPS from the opvABON strain did not change upon phage challenge (Fig 3D).
Confirmation that challenge of the wild type with P22, 9NA, and Det7 selected opvABON S. enterica cells was obtained by flow cytometry analysis (Fig 4). Expression of opvAB was monitored using a green fluorescent protein (gfp) fusion constructed downstream opvB (so that the strain remains OpvAB+). In the absence of phage, most S. enterica cells expressed opvAB at low levels; however, a small subpopulation that expressed opvAB at high levels was also detected. Phage challenge yielded mostly S. enterica cells with high levels of opvAB expression. These observations provide additional evidence that phages P22, 9NA, and Det7 kill the opvABOFF subpopulation, and that opvABON cells overtake the culture.
Fig. 4. Flow cytometry analysis of opvABOFF and opvABON subpopulations. 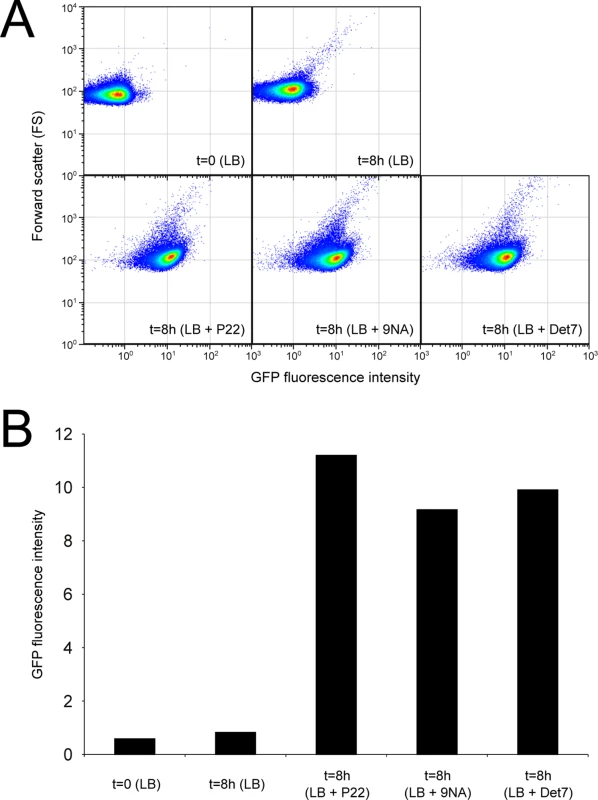
A. GFP fluorescence distribution in an ATCC 14028 derivative carrying an opvB::gfp fusion before (t = 0) and after growth in LB, LB + P22, LB + 9NA, and LB + Det7 (t = 8h). Data are represented by a dot plot (forward scatter [cellular size] versus fluorescence intensity [opvB::gfp expression]). All data were collected for 100,000 events per sample. B. Medians of GFP fluorescence from the same experiments. Reversibility of OpvAB-mediated bacteriophage resistance
If the above model was correct, we reasoned, cessation of phage challenge should permit resuscitation of a phage-sensitive subpopulation as a consequence of opvAB phase variation. This prediction was tested by isolating single colonies from cultures in LB + phage. After removal of phage by streaking on green plates, individual isolates were cultured in LB and re-challenged with P22, 9NA, and Det7 (≥ 20 isolates for each phage). All were phage-sensitive and their LPS profile was identical to that obtained before phage challenge. Representative examples are shown in Fig 5. Unlike the wild type, individual isolates of the ΔopvAB strain remained phage-resistant after single colony isolation and were considered mutants (see below).
Fig. 5. Reversibility of the phage-resistant phenotype in the wild type strain. 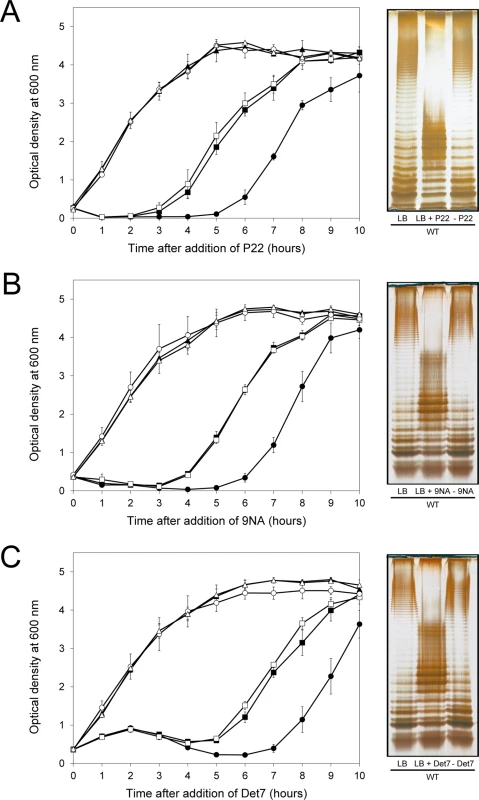
Left: Growth of the wild type strain (blue squares), an opvABON strain (green triangles) and a ΔopvAB strain (red circles) in LB + P22 (A), LB + 9NA (B), and LB + Det7 (C). The same symbols in different colors (wild type in purple, opvABON in yellow, ΔopvAB in brown) indicate phage-resistant isolates which were re-challenged. Values are averages and standard deviations from ≥ 3 independent experiments. Right: LPS profiles of the wild type strain in LB (left), LB + phage (center) and of an isolate that had survived phage challenge, subsequently grown in LB (right). LPS profiles were obtained by electrophoresis and silver staining. Mutational bacteriophage resistance in the absence of OpvAB
Challenge of a ΔopvAB strain with phages P22, 9NA, and Det7 prevented growth for 5–6 h, and growth resumed afterwards (Figs 3 and 5). To investigate the cause(s) of phage resistance in the absence of OpvAB, individual colonies were isolated from stationary cultures of a ΔopvAB strain in LB + P22, LB + 9NA, and LB + Det7. Phage was removed by streaking on green plates. Independent isolates (each from a different culture) were then tested for phage resistance. Sixty seven out of 72 independent isolates turned out to be phage-resistant, thus confirming that they were mutants. Analysis of LPS in independent phage-resistant mutants revealed that a large fraction of such mutants displayed visible LPS anomalies (Fig 6). The few mutant isolates (5/67) that did not show LPS alterations may have LPS alterations that cannot be detected in gels or carry mutations that confer phage resistance by mechanisms unrelated to the LPS. Whatever the case, these experiments support the conclusion that resistance of S. enterica to phages P22, 9NA, and Det7 in the absence of OpvAB is mutational.
Fig. 6. Mutational resistance to bacteriophage in a ΔopvAB strain. 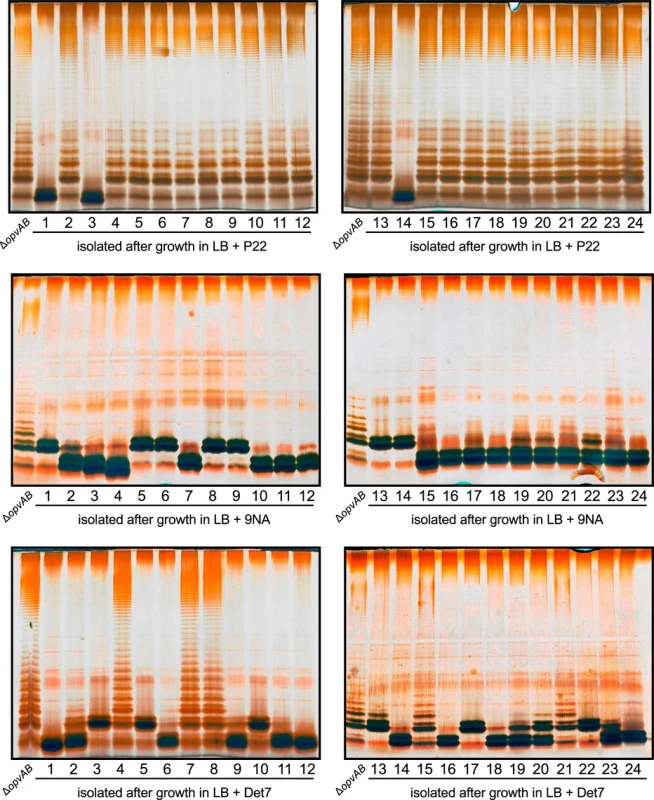
Top. LPS profiles of a ΔopvAB strain (first lane from the left on each gel) and 24 independent P22-resistant derivatives, as observed by electrophoresis and silver staining. Middle. LPS profiles of a ΔopvAB strain and 24 independent 9NA-resistant derivatives. Bottom. LPS profiles of a ΔopvAB strain and 24 independent Det7-resistant derivatives. To determine whether isolates resistant to one phage were also resistant to other phages that target the O-antigen, cross-resistance was tested by growth in LB upon phage inoculation. Sixty seven mutants (24 P22-resistant, 24 9NA-resistant, and 19 Det7-resistant) were tested (S1 Table). The main conclusions from these experiments were as follows:
Major alteration of LPS conferred resistance to all three phages.
More subtle LPS alteration (as observed in 16/24 P22-resistant mutants) conferred incomplete resistance to 9NA but did not confer resistance to Det7.
Effect of opvAB expression on Salmonella enterica virulence
Because the LPS plays roles in the interaction between S. enterica and the animal host [34,46,47], we tested whether OpvAB-mediated alteration of O-antigen chain length affected Salmonella virulence. For this purpose, competitive indexes (CI's) [48] were calculated in the following experiments: (i) oral and intraperitoneal inoculation of BALB/c mice; (ii) infection of mouse macrophages in vitro; and (iii) exposure to guinea pig serum, an assay that provides reductionist assessment of the capacity of the pathogen to survive the bactericidal activity of complement [47]. As controls, CI's were also calculated in LB (Table 1). In all virulence assays, the CI of the opvABON strain was found to be lower than those of the wild type and the ΔopvAB strain. Because the wild type, the opvABON strain and the ΔopvAB strain show similar or identical growth rates in LB, the conclusion from these experiments was that expression of opvAB reduces Salmonella virulence (Table 1).
Tab. 1. Competitive indexes of opvABON and ΔopvAB strains of S. enterica. 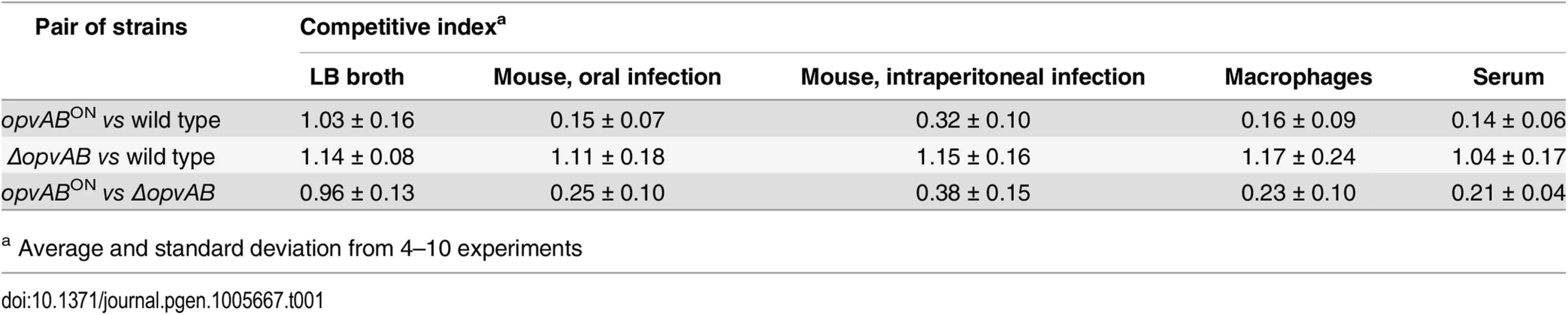
a Average and standard deviation from 4–10 experiments Discussion
A tradeoff is established whenever the adaptive capacity of an organism is increased at the expense of lowering the fitness conferred by specific phenotypic traits [49]. Tradeoffs have been mainly studied in sexually reproducing organisms but they occur also in microbes [50,51,52,53]. In pathogens, for instance, acquisition of mutational resistance to antimicrobial compounds often affects fitness [54,55], and may require loss of virulence as a payoff [56]. Bacteriophage resistance has been also shown to impair virulence in a variety of bacterial pathogens [57].
In this study, we describe a tradeoff that confers bacteriophage resistance at the expense of reducing virulence in the human pathogen Salmonella enterica. This tradeoff is however unusual because phage resistance is not mutational but epigenetic, and because the phage-resistant, avirulent phenotype is reversible.
The opvAB operon is present in most Salmonella serovars (S2 Table). Its products are inner membrane proteins that form intertwining ribbons (Fig 1) reminiscent of those formed by the LPS in the outer membrane [33]. Synthesis of OpvA and OpvB causes a decrease of long O-antigen chains and an increase of short O-antigen chains in the LPS (Fig 2; see also [32]). Genetic evidence presented in Fig 2 suggests that OpvA may prevent the formation of normal O-antigen, allowing OpvB to compete with the WzzST modal length regulator. A similar phenomenon occurs in Pseudomonas aeruginosa, where the Iap transmembrane peptide encoded by bacteriophage D3 disrupts endogenous O-antigen biosynthesis allowing a phage-encoded O-antigen polymerase to produce a different O-antigen [58]. OpvB confers a predominant modal length of 3–8 units, while the wild type LPS shows modal lenghts of 16–35 units and of >100 units (Fig 2; see also [32]). As a consequence of the dramatic change in LPS structure caused by opvAB expression, S. enterica becomes resistant to bacteriophages 9NA, Det7, and P22 (Figs 3 and 4), an observation consistent with the fact that the O-antigen is the bacterial surface receptor used by these bacteriophages [39,59].
Expression of opvAB undergoes phase variation under the control of DNA adenine methylation and the transcriptional regulator OxyR [32]. Because opvAB phase variation is skewed towards the OFF state [32], S. enterica populations contain a major subpopulation of opvABOFF (phage-sensitive) cells and a minor subpopulation of opvABON (phage-resistant) cells. In the presence of a bacteriophage that targets the O-antigen, the opvABOFF subpopulation disappears and the opvABON subpopulation is selected (Figs 3 and 4). Hence, the existence of a small subpopulation of phage-resistant cells preadapts S. enterica to survive phage challenge. In OpvAB−S. enterica, acquisition of phage resistance is mutational only, and a frequent mechanism is alteration of LPS structure (Fig 6). Because the LPS plays major roles in bacterial physiology including resistance to environmental injuries and host-pathogen interaction [60], opvAB phase variation may have selective value by providing S. enterica with a non-mutational, reversible mechanism of phage resistance. This mechanism offers the additional advantage of protecting Salmonella from multiple phages, perhaps from all phages that bind the O-antigen (note that the phages used in this study belong to three different families: Podoviridae, Siphoviridae, and Myoviridae).
Acquisition of phage resistance in opvABON cells requires a payoff: reduced virulence in both the mouse model and in vitro virulence assays (Table 1). In a phage-free environment, this payoff may be irrelevant because the avirulent subpopulation is minor as a consequence of skewed switching of opvAB toward the OFF state: 4 x 10−2 for ON→OFF switching vs 6 x 10−5 for OFF→ON switching [32]. In other words, only 1/1,000 S. enterica cells can be expected to be avirulent in a phage-free environment. The virulence payoff is therefore enforced in the presence of phage only, and its adaptive value may be obvious as it permits survival. On the other hand, the fitness cost of OpvAB-mediated phage resistance can be expected to be temporary because phase variation permits resuscitation of the virulent opvABOFF subpopulation as soon as phage challenge ceases (Fig 5). Resuscitation may actually be rapid as a consequence of skewed switching towards the opvABOFF state.
Phase variation systems that contribute to bacteriophage resistance have been described previously. For instance, certain restriction-modification systems show phase-variable expression [31]. However, protection by restriction-modification systems can be expected to be incomplete as only a fraction of infecting phage genomes are modified [61]. Phase variation can also confer phage resistance by preventing infection, and an interesting example is the gtrABC1 cluster which protects S. enterica against the T5-like phage SPC35 [28]. Although the receptor of SPC35 is the BtuB vitamin transporter, GtrABC-mediated glycosylation of the LPS O-antigen may reduce SPC35 adsorption by an indirect mechanism [28]. In Haemophilus influenzae, phase-variable resistance to bacteriophage HP1c1 may involve changes in LPS [29]. Because these studies did not investigate the impact of phase variation on bacterial fitness, it remains unknown whether the tradeoff associated with opvAB phase variation is unusual or commonplace. However, if one considers that envelope structures play multiple roles in bacterial physiology aside from serving as phage receptors, it is tempting to predict that phase-variable bacteriophage resistance may frequently involve fitness costs. Whatever the payoff, however, phase-variable resistance may have a crucial advantage over mutation by creating phenotypic heterogeneity in a reversible manner.
Methods
Bacterial strains
Strains of Salmonella enterica used in this study (Table 2) belong to serovar Typhimurium, and derive from the mouse-virulent strain ATCC 14028. For simplicity, S. enterica serovar Typhimurium is routinely abbreviated as S. enterica. For the construction of strain SV7643, a fragment containing the promoterless mCherry gene and the kanamycin resistance cassette was PCR-amplified from pDOC-R, an mCherry-containing derivative of plasmid pDOC [62] using primers HindIII-opvB-mCherry-5 and NdeI-opvB-mCherry-3. The construct was integrated into the chromosome of S. enterica using the Lambda Red recombination system [63]. For the construction of strains SV5675, SV6786, SV6791, and SV8020, targeted gene disruption was achieved using plasmid pKD13 [63] and oligonucleotides listed in S3 Table: wzzB5-PS4 + wzzB3-PS1 for wzzST disruption, fepE5-PS4 + fepE3-PS1 for wzzfepE disruption, STM2209-PS4tris + STM2209-PS1 for opvA disruption, and STM2208-PS4 + STM2208-PS1 for opvB disruption. The kanamycin resistance cassettes were then excised by recombination with plasmid pCP20 [63]. For the construction of strain SV6727, a fragment containing the promoterless green fluorescent protein (gfp) gene and the chloramphenicol resistance cassette was PCR-amplified from pZEP07 [64] using primers STM2208stop-GFP-5 and STM2208stop-GFP-3. The fragment was integrated into the chromosome of S. enterica using the Lambda Red recombination system [63]. An opvB::gfp transcriptional fusion was formed downstream of the opvB stop codon, and the strain remained OpvAB+. For the construction of strains SV7645, SV8117, and SV8118, plasmid pKD46 was introduced in SV6401, and the PCR products used for construction of strains SV7643, SV8020 and SV5675 were integrated into the chromosome of SV6401 using the Lambda Red recombination system [63].
Tab. 2. Strain list. 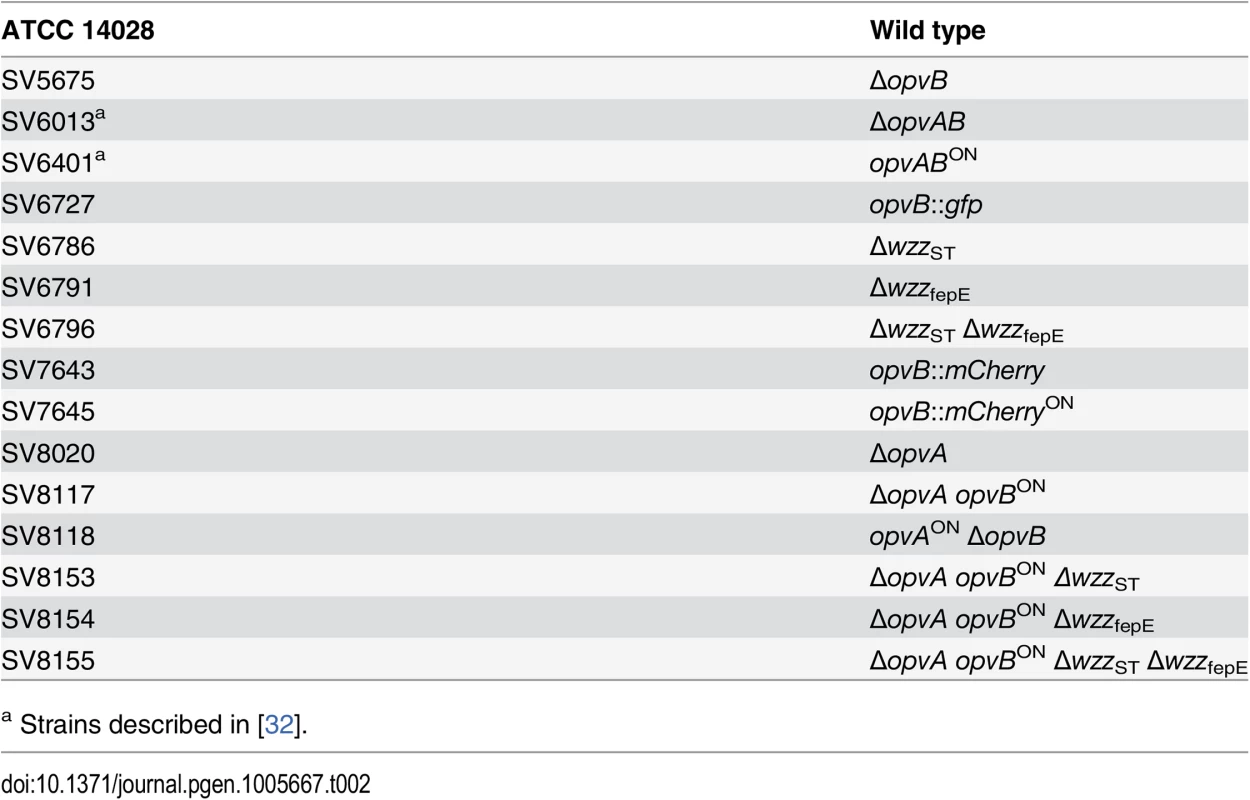
a Strains described in [32]. Culture media
Bertani's lysogeny broth (LB) was used as standard liquid medium. Solid LB contained agar at 1.5% final concentration. Green plates [65] contained methyl blue (Sigma-Aldrich, St. Louis, MO) instead of aniline blue. Antibiotics were used at the concentrations described previously [66].
Bacteriophages
Bacteriophages 9NA [41,42] and Det7 [43] were kindly provided by Sherwood Casjens, University of Utah, Salt Lake City. Bacteriophage P22 H5 is a virulent derivative of bacteriophage P22 that carries a mutation in the c2 gene [45], and was kindly provided by John R. Roth, University of California, Davis. For simplicity, P22 H5 is abbreviated as P22 throughout the text.
Construction of plasmid pIZ2011
A DNA fragment containing opvA and the native opvAB promoter was PCR-amplified using primers KpnI-opvA-plasmidoGFP-5 and KpnI-opvA-plasmidoGFP-3 (S3 Table). The amplification product was cloned into pDOC-R [62]. The resulting plasmid produces an OpvA-mCherry fusion protein.
Fluorescence microscopy
Bacterial cells from 1.5 ml of an exponential culture in LB at 37°C (OD600 ~0.15) were collected by centrifugation, washed in phosphate saline buffer (PBS), and resuspended in 1 ml of the same buffer. Cells were fixed in 4% formaldehyde solution and incubated at room temperature for 30 minutes. Finally, cells were washed, resuspended in PBS buffer, and stored at 4°C. Images were obtained by using an Olympus IX-70 Delta Vision fluorescence microscope equipped with a 100X UPLS Apo objective. Pictures were taken using a CoolSNAP HQ/ICX285 camera and analyzed using ImageJ software (Wayne Rasband, Research Services Branch, National Institute of Mental Health, MD, USA). Z-stacks (optical sections separated by 0.2 μm) of mCherry fluorescence were taken with the same microscope. Maximal intensity projections are shown.
Electrophoretic visualization of LPS profiles
To investigate LPS profiles, bacterial cultures were grown in LB overnight. Bacterial cells were harvested and washed with 0.9% NaCl. The O.D.600 of the washed bacterial suspension was measured to calculate cell concentration. A bacterial mass containing about 3 x 108 cells was pelleted by centrifugation. Treatments applied to the bacterial pellet, electrophoresis of crude bacterial extracts, and silver staining procedures were performed as described by Buendia-Claveria et al. [67].
Flow cytometry
Bacterial cultures were grown at 37°C in LB or LB + phage (P22, 9NA, or Det7) until exponential (OD600 ~0.3) or stationary phase (OD600 ~4). Cells were then diluted in PBS. Data acquisition and analysis were performed using a Cytomics FC500-MPL cytometer (Beckman Coulter, Brea, CA). Data were collected for 100,000 events per sample, and analyzed with CXP and FlowJo 8.7 software.
Bacteriophage challenge
Overnight cultures were diluted 1 : 100 in 3 ml LB and grown in aeration by shaking at 37°C until they reached an optical density OD600 ~0.3. One hundred μl of a bacteriophage lysate (P22 H5, 9NA, or Det7) were added (M.O.I. ≥10), and OD600 was subsequently measured at 1 h intervals.
Virulence assays
Eight-week-old female BALB/c mice (Charles River Laboratories, Santa Perpetua de Mogoda, Spain) were inoculated with pairwise combinations of the wild type, an opvABON strain, and a ΔopvAB strain at a 1 : 1 ratio. Bacterial cultures were previously grown overnight at 37°C in LB without shaking. Oral inoculation was performed by feeding the mice with 25 μl of PBS containing 0.1% lactose and 108 bacterial colony-forming units (CFU). Intraperitoneal inoculation was performed with 104 CFU in 200 μl of PBS. Bacteria were recovered from the spleen and the liver of infected mice at 2 days post-infection (intraperitoneal challenge) or 5 days post-infection (oral challenge). A competitive index (CI) was calculated as described elsewhere [48]. To permit strain discrimination, ATCC 14208 was tagged with trg::MudJ (Kmr), an allele that is neutral for virulence [68]. When necessary, cross-streaking on green plates with P22 H5 was used to discriminate phage-resistant isolates [65]. Infection of cultured J774 mouse macrophages, inoculation of guinea pig serum (Sigma-Aldrich), and calculation of competitive indexes in vitro followed previously described protocols [68]. The Student's t test was used to determine whether the CI's were significant.
Ethics statement
Animal research adhered to the principles mandatory in the European Union, as established in the Legislative Act 86/609 CEE (November 24, 1986) and followed the specific protocols established by the Royal Decree 1201/2005 of the Government of Spain (October 10, 2005). The protocols employed in the study were reviewed by the Comité Ético de Experimentación of the Consejo Superior de Investigaciones Científicas (CSIC), and were approved by the Consejería de Medio Ambiente, Comunidad de Madrid, Spain, on December 12, 2014 (permit number PROEX 257/14).
Supporting Information
Zdroje
1. Shapiro L (1976) Differentiation in the Caulobacter cell cycle. Annu Rev Microbiol 30 : 377–407. 185940
2. Kaiser D (1986) Control of multicellular development: Dictyostelium and Myxococcus. Annu Rev Genet 20 : 539–566. 3028248
3. Stragier P, Losick R (1996) Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet 30 : 297–241. 8982457
4. Andrewes FW (1922) Studies in group agglutination. I. The Salmonella group and its antigenic structure. J Path Bact 25 : 505–524.
5. Legroux R, Magrou J (1920) État organisé des colonies bactériennes. Ann Inst Pasteur (Paris) 34 : 417–431.
6. Shapiro JA, Higgins NP (1989) Differential activity of a transposable element in Escherichia coli colonies. J Bacteriol 171 : 5975–5986. 2553666
7. Shapiro JA (1998) Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol 52 : 81–104. 9891794
8. Zgur-Bertok D (2007) Phenotypic heterogeneity in bacterial populations. Acta Agric Slovenica 90 : 17–24.
9. Dhar N, McKinney JD (2007) Microbial phenotypic heterogeneity and antibiotic tolerance. Curr Opin Microbiol 10 : 30–38. 17215163
10. Davidson CJ, Surette MG (2008) Individuality in bacteria. Annu Rev Genet 42 : 253–268. doi: 10.1146/annurev.genet.42.110807.091601 18652543
11. Veening JW, Smits WK, Kuipers OP (2008) Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62 : 193–210. doi: 10.1146/annurev.micro.62.081307.163002 18537474
12. Casadesus J, Low DA (2013) Programmed heterogeneity: epigenetic mechanisms in bacteria. J Biol Chem 288 : 13929–13935. doi: 10.1074/jbc.R113.472274 23592777
13. Thattai M, van Oudenaarden A (2004) Stochastic gene expression in fluctuating environments. Genetics 167 : 523–530. 15166174
14. Kussell E, Leibler S (2005) Phenotypic diversity, population growth, and information in fluctuating environments. Science 309 : 2075–2078. 16123265
15. Wolf DM, Vazirani VV, Arkin AP (2005) Diversity in times of adversity: probabilistic strategies in microbial survival games. J Theor Biol 234 : 227–253. 15757681
16. Hernandez SB, Cota I, Ducret A, Aussel L, Casadesus J (2012) Adaptation and preadaptation of Salmonella enterica to bile. PLoS Genet 8: e1002459. doi: 10.1371/journal.pgen.1002459 22275872
17. Sanchez-Romero MA, Casadesus J (2014) Contribution of phenotypic heterogeneity to adaptive antibiotic resistance. Proc Natl Acad Sci U S A 111 : 355–360. doi: 10.1073/pnas.1316084111 24351930
18. Ni M, Decrulle AL, Fontaine F, Demarez A, Taddei F, et al. (2012) Pre-disposition and epigenetics govern variation in bacterial survival upon stress. PLoS Genet 8: e1003148. doi: 10.1371/journal.pgen.1003148 23284305
19. Silverman M, Zieg J, Hilmen M, Simon M (1979) Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc Natl Acad Sci U S A 76 : 391–395. 370828
20. Moxon ER, Rainey PB, Nowak MA, Lenski RE (1994) Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol 4 : 24–33. 7922307
21. Moxon R, Bayliss C, Hood D (2006) Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu Rev Genet 40 : 307–333. 17094739
22. Dubnau D, Losick R (2006) Bistability in bacteria. Mol Microbiol 61 : 564–572. 16879639
23. Hernday A, Krabbe M, Braaten B, Low D (2002) Self-perpetuating epigenetic pili switches in bacteria. Proc Natl Acad Sci U S A 99 : 16470–16476. 12202745
24. Casadesus J, Low D (2006) Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev 70 : 830–856. 16959970
25. van der Woude MW (2011) Phase variation: how to create and coordinate population diversity. Curr Opin Microbiol 14 : 205–211. doi: 10.1016/j.mib.2011.01.002 21292543
26. van der Woude MW, Baumler AJ (2004) Phase and antigenic variation in bacteria. Clin Microbiol Rev 17 : 581–611, table of contents. 15258095
27. van der Woude MW (2006) Re-examining the role and random nature of phase variation. FEMS Microbiol Lett 254 : 190–197. 16445745
28. Kim M, Ryu S (2012) Spontaneous and transient defence against bacteriophage by phase-variable glucosylation of O-antigen in Salmonella enterica serovar Typhimurium. Mol Microbiol 86 : 411–425. doi: 10.1111/j.1365-2958.2012.08202.x 22928771
29. Zaleski P, Wojciechowski M, Piekarowicz A (2005) The role of Dam methylation in phase variation of Haemophilus influenzae genes involved in defence against phage infection. Microbiology 151 : 3361–3369. 16207918
30. Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, Jennings MP (2005) The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc Natl Acad Sci U S A 102 : 5547–5551. 15802471
31. Hoskisson PA, Smith MC (2007) Hypervariation and phase variation in the bacteriophage 'resistome'. Curr Opin Microbiol 10 : 396–400. 17719266
32. Cota I, Blanc-Potard AB, Casadesus J (2012) STM2209-STM2208 (opvAB): a phase variation locus of Salmonella enterica involved in control of O-antigen chain length. PLoS One 7: e36863. doi: 10.1371/journal.pone.0036863 22606300
33. Ghosh AS, Young KD (2005) Helical disposition of proteins and lipopolysaccharide in the outer membrane of Escherichia coli. J Bacteriol 187 : 1913–1922. 15743937
34. Murray GL, Attridge SR, Morona R (2003) Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol Microbiol 47 : 1395–1406. 12603743
35. Goldman RC, Hunt F (1990) Mechanism of O-antigen distribution in lipopolysaccharide. J Bacteriol 172 : 5352–5359. 1697578
36. Bastin DA, Stevenson G, Brown PK, Haase A, Reeves PR (1993) Repeat unit polysaccharides of bacteria: a model for polymerization resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol Microbiol 7 : 725–734. 7682279
37. Daniels C, Morona R (1999) Analysis of Shigella flexneri Wzz (Rol) function by mutagenesis and cross-linking: Wzz is able to oligomerize. Mol Microbiol 34 : 181–194. 10540296
38. Batchelor RA, Alifano P, Biffali E, Hull SI, Hull RA (1992) Nucleotide sequences of the genes regulating O-polysaccharide antigen chain length (rol) from Escherichia coli and Salmonella typhimurium: protein homology and functional complementation. J Bacteriol 174 : 5228–5236. 1379582
39. Lindberg AA (1973) Bacteriophage receptors. Annu Rev Microbiol 27 : 205–241. 4584686
40. Kintz E, Davies MR, Hammarlof DL, Canals R, Hinton JC, et al. (2015) A BTP1 prophage gene present in invasive non-typhoidal Salmonella determines composition and length of the O-antigen of the lipopolysaccharide. Mol Microbiol 96 : 263–275. doi: 10.1111/mmi.12933 25586744
41. Wilkinson RG, Gemski P Jr., Stocker BA (1972) Non-smooth mutants of Salmonella typhimurium: differentiation by phage sensitivity and genetic mapping. J Gen Microbiol 70 : 527–554. 4556257
42. Casjens SR, Leavitt JC, Hatfull GF, Hendrix RW (2014) Genome sequence of Salmonella phage 9NA. Genome Announc 2: e00531–14 doi: 10.1128/genomeA.00531-14 25146133
43. Walter M, Fiedler C, Grassl R, Biebl M, Rachel R, et al. (2008) Structure of the receptor-binding protein of bacteriophage det7: a podoviral tail spike in a myovirus. J Virol 82 : 2265–2273. 18077713
44. Casjens SR, Jacobs-Sera D, Hatfull GF, Hendrix RW (2015) Genome sequence of Salmonella enterica phage Det7. Genome Announc 3: e00279–00215. doi: 10.1128/genomeA.00279-15 25953168
45. Smith HO, Levine M (1964) Two sequential repressions of DNA synthesis in the establishment of lysogeny by phage P22 and Its mutants. Proc Natl Acad Sci U S A 52 : 356–363. 14206603
46. Raetz CR, Whitfield C (2002) Lipopolysaccharide endotoxins. Annu Rev Biochem 71 : 635–700. 12045108
47. Bravo D, Silva C, Carter JA, Hoare A, Alvarez SA, et al. (2008) Growth-phase regulation of lipopolysaccharide O-antigen chain length influences serum resistance in serovars of Salmonella. J Med Microbiol 57 : 938–946. doi: 10.1099/jmm.0.47848-0 18628492
48. Beuzon CR, Holden DW (2001) Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect 3 : 1345–1352. 11755424
49. Williams GC (1966) Natural selection, the costs of reproduction, and a refinement of Lack's principle. Am Nat 100 : 687–690.
50. Nystrom T (2004) Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol 54 : 855–862. 15522072
51. Shoval O, Sheftel H, Shinar G, Hart Y, Ramote O, et al. (2012) Evolutionary trade-offs, Pareto optimality, and the geometry of phenotype space. Science 336 : 1157–1160. doi: 10.1126/science.1217405 22539553
52. Bailly-Bechet M, Benecke A, Hardt WD, Lanza V, Sturm A, et al. (2011) An externally modulated, noise-driven switch for the regulation of SPI1 in Salmonella enterica serovar Typhimurium. J Math Biol 63 : 637–662. doi: 10.1007/s00285-010-0385-1 21107576
53. Ferenci T, Spira B (2007) Variation in stress responses within a bacterial species and the indirect costs of stress resistance. Ann N Y Acad Sci 1113 : 105–113. 17483210
54. Andersson DI, Hughes D (2010) Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8 : 260–271. doi: 10.1038/nrmicro2319 20208551
55. De Paepe M, Gaboriau-Routhiau V, Rainteau D, Rakotobe S, Taddei F, et al. (2011) Trade-off between bile resistance and nutritional competence drives Escherichia coli diversification in the mouse gut. PLoS Genet 7: e1002107. doi: 10.1371/journal.pgen.1002107 21698140
56. Vincent BM, Lancaster AK, Scherz-Shouval R, Whitesell L, Lindquist S (2013) Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS Biol 11: e1001692. doi: 10.1371/journal.pbio.1001692 24204207
57. Leon M, Bastias R (2015) Virulence reduction in bacteriophage resistant bacteria. Front Microbiol 6 : 343. doi: 10.3389/fmicb.2015.00343 25954266
58. Taylor VL, Udaskin ML, Islam ST, Lam JS (2013) The D3 bacteriophage alpha-polymerase inhibitor (Iap) peptide disrupts O-antigen biosynthesis through mimicry of the chain length regulator Wzz in Pseudomonas aeruginosa. J Bacteriol 195 : 4735–4741. doi: 10.1128/JB.00903-13 23955007
59. Susskind MM, Botstein D (1978) Molecular genetics of bacteriophage P22. Microbiol Rev 42 : 385–413. 353481
60. Whitfield C, Trent MS (2014) Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem 83 : 99–128. doi: 10.1146/annurev-biochem-060713-035600 24580642
61. Wilson GG, Murray NE (1991) Restriction and modification systems. Annu Rev Genet 25 : 585–627. 1812816
62. Lee DJ, Bingle LE, Heurlier K, Pallen MJ, Penn CW, et al. (2009) Gene doctoring: a method for recombineering in laboratory and pathogenic Escherichia coli strains. BMC Microbiol 9 : 252. doi: 10.1186/1471-2180-9-252 20003185
63. Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97 : 6640–6645. 10829079
64. Hautefort I, Proenca MJ, Hinton JC (2003) Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl Environ Microbiol 69 : 7480–7491. 14660401
65. Chan RK, Botstein D, Watanabe T, Ogata Y (1972) Specialized transduction of tetracycline by phage P22 in Salmonella typhimurium. II. Properties of a high frequency transducing lysate. Virology 50 : 883–898. 4565618
66. Torreblanca J, Casadesús J (1996) DNA adenine methylase mutants of Salmonella typhimurium and a novel Dam-regulated locus. Genetics 144 : 15–26. 8878670
67. Buendia-Claveria AM, Moussaid A, Ollero FJ, Vinardell JM, Torres A, et al. (2003) A purL mutant of Sinorhizobium fredii HH103 is symbiotically defective and altered in its lipopolysaccharide. Microbiology 149 : 1807–1818. 12855732
68. Segura I, Casadesus J, Ramos-Morales F (2004) Use of mixed infections to study cell invasion and intracellular proliferation of Salmonella enterica in eukaryotic cell cultures. J Microbiol Methods 56 : 83–91. 14706753
Štítky
Genetika Reprodukční medicína
Článek A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin TransporterČlánek Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis inČlánek Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 11
-
Všechny články tohoto čísla
- Agricultural Genomics: Commercial Applications Bring Increased Basic Research Power
- Ernst Rüdin’s Unpublished 1922-1925 Study “Inheritance of Manic-Depressive Insanity”: Genetic Research Findings Subordinated to Eugenic Ideology
- Convergent Evolution During Local Adaptation to Patchy Landscapes
- The Locus Controls Age at Maturity in Wild and Domesticated Atlantic Salmon ( L.) Males
- A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin Transporter
- Absence of Maternal Methylation in Biparental Hydatidiform Moles from Women with Maternal-Effect Mutations Reveals Widespread Placenta-Specific Imprinting
- Calibrating the Human Mutation Rate via Ancestral Recombination Density in Diploid Genomes
- Anaplastic Lymphoma Kinase Acts in the Mushroom Body to Negatively Regulate Sleep
- Connecting Replication and Repair: YoaA, a Helicase-Related Protein, Promotes Azidothymidine Tolerance through Association with Chi, an Accessory Clamp Loader Protein
- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Mosaic and Intronic Mutations in Explain the Majority of TSC Patients with No Mutation Identified by Conventional Testing
- Members of the Epistasis Group Contribute to Mitochondrial Homologous Recombination and Double-Strand Break Repair in
- QTL Mapping of Sex Determination Loci Supports an Ancient Pathway in Ants and Honey Bees
- Genetic Interactions Implicating Postreplicative Repair in Okazaki Fragment Processing
- Genomics of Cancer and a New Era for Cancer Prevention
- Adaptation to High Ethanol Reveals Complex Evolutionary Pathways
- Dynamics of Transcription Factor Binding Site Evolution
- Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis in
- Enhancer Runaway and the Evolution of Diploid Gene Expression
- Cattle Sex-Specific Recombination and Genetic Control from a Large Pedigree Analysis
- Drosophila Mutants Model Cornelia de Lange Syndrome in Growth and Behavior
- Pleiotropic Effects of Immune Responses Explain Variation in the Prevalence of Fibroproliferative Diseases
- Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape
- Tissue-Specific Effects of Reduced β-catenin Expression on Mutation-Instigated Tumorigenesis in Mouse Colon and Ovarian Epithelium
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Mapping of Craniofacial Traits in Outbred Mice Identifies Major Developmental Genes Involved in Shape Determination
- Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- DNA Repair Cofactors ATMIN and NBS1 Are Required to Suppress T Cell Activation
- Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
- Ernst Rüdin and the State of Science
- ABCs of Insect Resistance to Bt
- Epigenetic Control of O-Antigen Chain Length: A Tradeoff between Virulence and Bacteriophage Resistance
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
- The Fanconi Anemia Pathway Protects Genome Integrity from R-loops
- Controls Quantitative Variation in Maize Kernel Row Number
- Genome-Wide Association Study of Golden Retrievers Identifies Germ-Line Risk Factors Predisposing to Mast Cell Tumours
- Insect Resistance to Toxin Cry2Ab Is Conferred by Mutations in an ABC Transporter Subfamily A Protein
- A Cytosine Methytransferase Modulates the Cell Envelope Stress Response in the Cholera Pathogen
- Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals
- The Multi-allelic Genetic Architecture of a Variance-Heterogeneity Locus for Molybdenum Concentration in Leaves Acts as a Source of Unexplained Additive Genetic Variance
- The lncRNA Controls Cryptococcal Morphological Transition
- Sae2 Function at DNA Double-Strand Breaks Is Bypassed by Dampening Tel1 or Rad53 Activity
- A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia,
- Ectodysplasin/NF-κB Promotes Mammary Cell Fate via Wnt/β-catenin Pathway
- The QTL within the Complex Involved in the Control of Tuberculosis Infection in Mice Is the Classical Class II Gene
- Identifying Loci Contributing to Natural Variation in Xenobiotic Resistance in
- Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by and Subsistence
- A Flexible, Efficient Binomial Mixed Model for Identifying Differential DNA Methylation in Bisulfite Sequencing Data
- Competition between Heterochromatic Loci Allows the Abundance of the Silencing Protein, Sir4, to Regulate Assembly of Heterochromatin
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



