-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaInhibition of Competence Development, Horizontal Gene Transfer and Virulence in by a Modified Competence Stimulating Peptide
Competence stimulating peptide (CSP) is a 17-amino acid peptide pheromone secreted by Streptococcus pneumoniae. Upon binding of CSP to its membrane-associated receptor kinase ComD, a cascade of signaling events is initiated, leading to activation of the competence regulon by the response regulator ComE. Genes encoding proteins that are involved in DNA uptake and transformation, as well as virulence, are upregulated. Previous studies have shown that disruption of key components in the competence regulon inhibits DNA transformation and attenuates virulence. Thus, synthetic analogues that competitively inhibit CSPs may serve as attractive drugs to control pneumococcal infection and to reduce horizontal gene transfer during infection. We performed amino acid substitutions on conserved amino acid residues of CSP1 in an effort to disable DNA transformation and to attenuate the virulence of S. pneumoniae. One of the mutated peptides, CSP1-E1A, inhibited development of competence in DNA transformation by outcompeting CSP1 in time and concentration-dependent manners. CSP1-E1A reduced the expression of pneumococcal virulence factors choline binding protein D (CbpD) and autolysin A (LytA) in vitro, and significantly reduced mouse mortality after lung infection. Furthermore, CSP1-E1A attenuated the acquisition of an antibiotic resistance gene and a capsule gene in vivo. Finally, we demonstrated that the strategy of using a peptide inhibitor is applicable to other CSP subtype, including CSP2. CSP1-E1A and CSP2-E1A were able to cross inhibit the induction of competence and DNA transformation in pneumococcal strains with incompatible ComD subtypes. These results demonstrate the applicability of generating competitive analogues of CSPs as drugs to control horizontal transfer of antibiotic resistance and virulence genes, and to attenuate virulence during infection by S. pneumoniae.
Published in the journal: . PLoS Pathog 7(9): e32767. doi:10.1371/journal.ppat.1002241
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002241Summary
Competence stimulating peptide (CSP) is a 17-amino acid peptide pheromone secreted by Streptococcus pneumoniae. Upon binding of CSP to its membrane-associated receptor kinase ComD, a cascade of signaling events is initiated, leading to activation of the competence regulon by the response regulator ComE. Genes encoding proteins that are involved in DNA uptake and transformation, as well as virulence, are upregulated. Previous studies have shown that disruption of key components in the competence regulon inhibits DNA transformation and attenuates virulence. Thus, synthetic analogues that competitively inhibit CSPs may serve as attractive drugs to control pneumococcal infection and to reduce horizontal gene transfer during infection. We performed amino acid substitutions on conserved amino acid residues of CSP1 in an effort to disable DNA transformation and to attenuate the virulence of S. pneumoniae. One of the mutated peptides, CSP1-E1A, inhibited development of competence in DNA transformation by outcompeting CSP1 in time and concentration-dependent manners. CSP1-E1A reduced the expression of pneumococcal virulence factors choline binding protein D (CbpD) and autolysin A (LytA) in vitro, and significantly reduced mouse mortality after lung infection. Furthermore, CSP1-E1A attenuated the acquisition of an antibiotic resistance gene and a capsule gene in vivo. Finally, we demonstrated that the strategy of using a peptide inhibitor is applicable to other CSP subtype, including CSP2. CSP1-E1A and CSP2-E1A were able to cross inhibit the induction of competence and DNA transformation in pneumococcal strains with incompatible ComD subtypes. These results demonstrate the applicability of generating competitive analogues of CSPs as drugs to control horizontal transfer of antibiotic resistance and virulence genes, and to attenuate virulence during infection by S. pneumoniae.
Introduction
Streptococcus pneumoniae is one of the most important pathogens that cause bacterial pneumonia, otitis media, meningitis and sepsis [1]–[3]. The incidence of antibiotic resistance in S. pneumoniae has increased dramatically in the recent decades [4]–[6]. The acquisition and spread of antibiotic resistance genes in S. pneumoniae is at least partly due to genetic transformation, which occurs when the bacteria enter the competent state [7]–[9]. The binding of competence stimulating peptide (CSP) to its membrane-associated histidine kinase receptor ComD initiates competence in S. pneumoniae [7]–[10]. Upon interacting with CSP, ComD phosphorylates the cognate transcriptional regulator ComE [11]–[13]. ComE then initiates the transcription of a set of 24 genes (early genes), including comX. As an alternative sigma factor, ComX positively regulates the transcription of 81 genes (late genes) in the competence regulon [14]–[15]. Some of these late genes encode effectors for DNA uptake and recombination [14]. DNA sequence analysis from 60 pneumococcal isolates predicts the existence of six distinct CSP subtypes [16], [17]. However, an overwhelming majority of S. pneumoniae strains produce two of these subtypes: CSP1 or CSP2 [17]. Furthermore, apart from CSP1 and CSP2, the ability of other CSP subtypes to induce DNA transformation has not been analyzed. There are two major corresponding variants of ComD, named ComD1 and ComD2 [18]. CSP1 and CSP2 share 50% amino acid identity. The major sequence variation between CSP1 and CSP2 occurs in the central region of these peptides, which confers receptor specificity [19]. In contrast, the first three amino acid residues of the N-terminus and the last two amino acid residues of the C-terminus are conserved between CSP1 and CSP2. Competence in pneumococcal strains with the ComD1 receptor could be induced more efficiently with the “compatible” CSP1. In contrast, ComD2 strains are more sensitive to induction by the “compatible” CSP2 [19].
Competence for DNA transformation plays a crucial role in the ability of S. pneumoniae to gain virulence and antibiotic resistance genes from other species. Importantly, in recent years, the competence regulon of S. pneumoniae has been shown to cross regulate virulence [20]–[24]. For example, Lau et al [20] reported that a loss of function in ComB, an accessory protein to the ComA ABC transporter required for the export of CSP, as well as a loss of function in the ComD histidine kinase, attenuate the ability of S. pneumoniae to cause pneumonia and bacteremia in mice. This report has been subsequently confirmed by other studies [21]–[23]. In addition, it has been shown that competence-mediated cell lysis may mediate the release of pneumolysin, as well as the cell wall component lipoteichoic acid (LTA), the former being an important virulence factor of S. pneumoniae [23], [24]. Microarray analysis has indicated that CSP induces the transcription of both virulence and stress responsive genes [14]. These above-mentioned studies indicate that activation of the competence regulon is not only essential for DNA uptake and transformation, but also important for virulence. Therefore, the competence regulon represents an attractive drug target to combat both pneumococcal infections and the spread of antibiotic resistance genes.
In this study, we performed amino acid substitutions in CSP1 and CSP2, and examined the ability of these CSP variants to inhibit the development of competence, horizontal gene transfer, and virulence of S. pneumoniae both in vitro and in mice. One of these modified peptides, CSP1-E1A, was able to competitively inhibit the development of competence for DNA transformation and expression of virulence gene in vitro. Importantly, CSP1-E1A was able to attenuate the virulence of S. pneumoniae during lung infection in mice, as well as inhibiting the ability of pneumococcus to acquire both an antibiotic resistance gene and a capsule gene during mouse models of acute pneumonia and bacteremia infections. Significantly, we demonstrated that the same amino acid substitution on CSP2 (CSP2-E1A) also inhibits CSP2-mediated competence development. Moreover, both CSP1-E1A and CSP2-E1A were capable of cross inhibiting the induction of competence regulon mediated by incompatible receptors, ComD2 and ComD1, respectively.
Results
The first and the third amino acid residues on the N terminus of CSP1 are critical for the induction of competence
Because amino acid residues in the central region of CSP1 are important for receptor specificity [19], we hypothesized that conserved amino acid residues on both the N-terminus and the C-terminus of CSP1 are important for its ability to induce competence. By amino acid substitutions or deletions of the first three amino acid residues on the N-terminus and of the last two amino acids on the C-terminus, we synthesized five CSP1 variants (Figure 1). The ability of these modified peptides to induce competence in S. pneumoniae wild-type strain D39 (Table 1) grown in Todd Hewitt Broth (THB) was compared to that of CSP1 by: (i) monitoring the transformation frequency of a rpsL gene that confers resistance to streptomycin [25], and (ii) monitoring the promoter activity of the competence specific sigma factor gene comX. Induction of ComX indicates that S. pneumoniae cells have entered a competent state. The promoter activity of comX was monitored by assaying the ß-galactosidase activity in D39pcomX::lacZ, a D39 derivative with a lacZ gene fused behind the comX promoter (Table 1) [26]. Deletion or substitution of either of the two lysine (K) residues on the C-terminus (CSP1-K16DK17D, CSP1-ΔK16ΔK17) did not alter the ability of these peptides to induce competence (Fig. 2A–2B). In contrast, substitution of the first amino acid (glutamate to alanine) (CSP1-E1A) severely reduced the ability of this peptide to induce comX (Fig. 2A) and DNA transformation (Fig. 2B). CSP1-E1A could only induce about 1% of comX promoter activity and > 3 logs lower DNA transformation when compared to CSP1. Similarly, alanine substitution of the third residue of the N terminus (CSP1-R3A) impaired its ability to induce both comX and DNA transformation (Fig. 2A–2B). In contrast, alanine substitution of the second amino acid residue on the N terminus (CSP1-M2A) did not alter its ability to induce both comX and DNA transformation (Fig. 2A–2B).
Fig. 1. Amino acid sequences of CSP1 and CSP2, and their analogues. 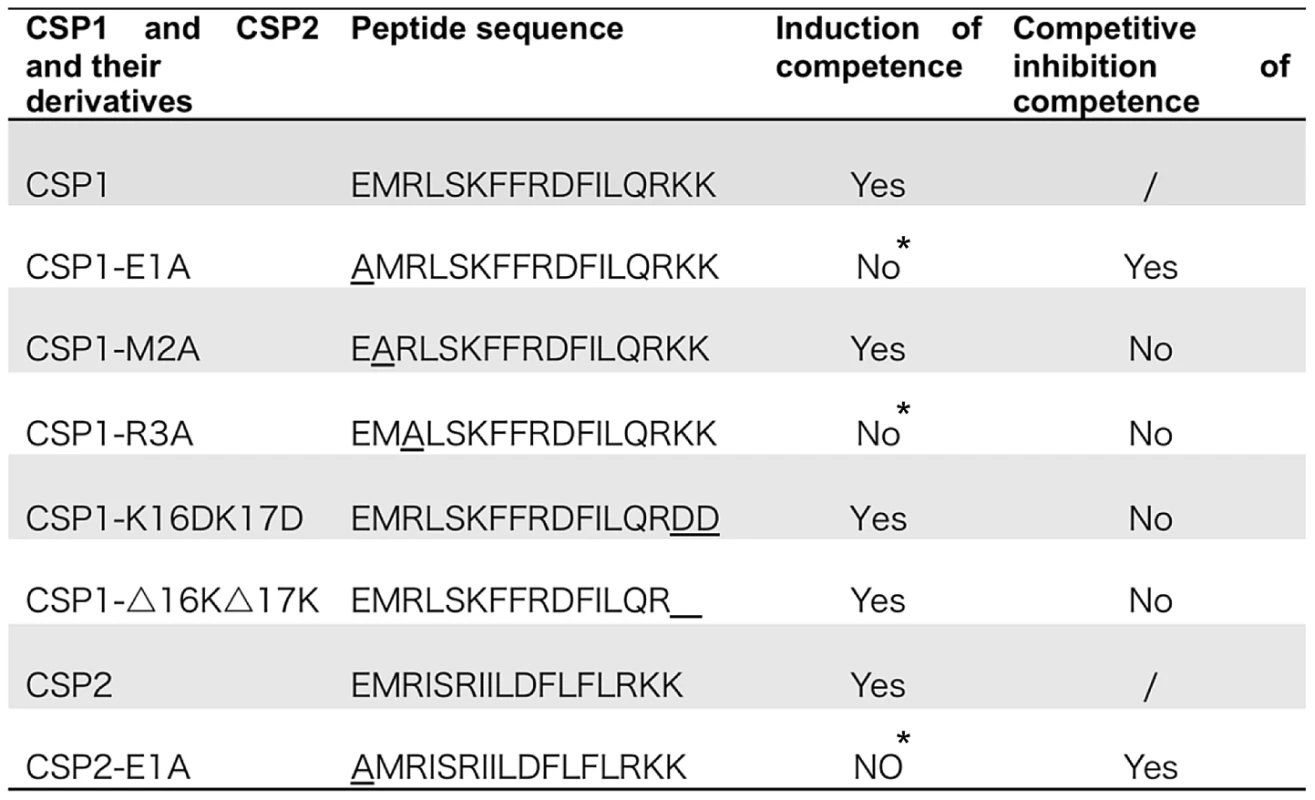
Modified CSPs were synthesized based on the sequence of CSP1 or CSP2 by amino acid substitutions or by deletion. The ability of each CSP analogue to induce competence and to competitively inhibit CSP1 or CSP2-mediated competence is presented. *No represents < 1% of CSP1 or CSP2 activity during the induction of competence. Fig. 2. The first and third amino acid residues of CSP1 are essential for induction of competence. 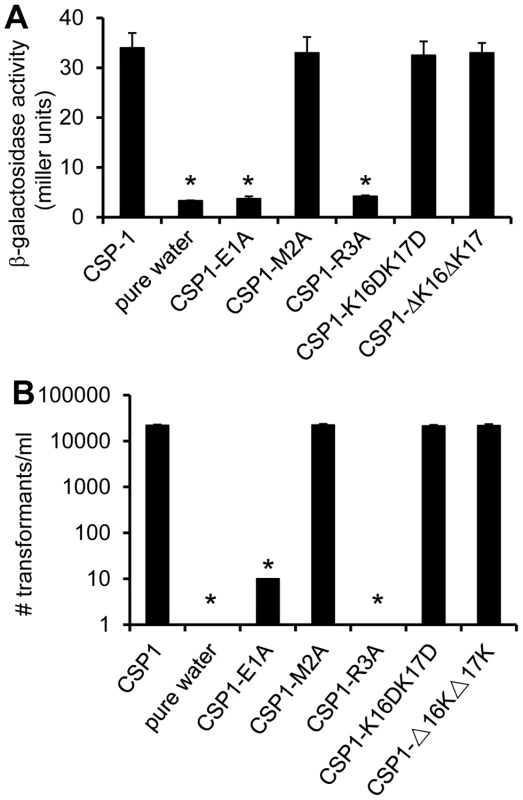
(A) D39pcomX::lacZ cells were incubated with CSP1 or with individual modified peptides at a final concentration of 100 ng/ml for 30 min. The ability of each peptide to induce competence regulon was measured by induction of the comX promoter in D39pcomX::lacZ. The amino acid substitution of 1st amino acid (CSP1-E1A) and 3rd amino acid (CSP1-R3A) in the N-terminus abolished the peptide's ability to induce the expression of comX. (B) D39 cells were exposed to 100 ng/ml of CSP1 or individual modified peptides. DNA transformation was performed using the rpsL gene. Transformants were selected on THB agar containing 100 µg/ml streptomycin. Experiments were performed in triplicates and repeated three times. The means ± SD of one typical experiment are shown. *p<0.01 when compared against CSP1-treated cells. Tab. 1. <i>S. pneumoniae</i> strains used in this study. 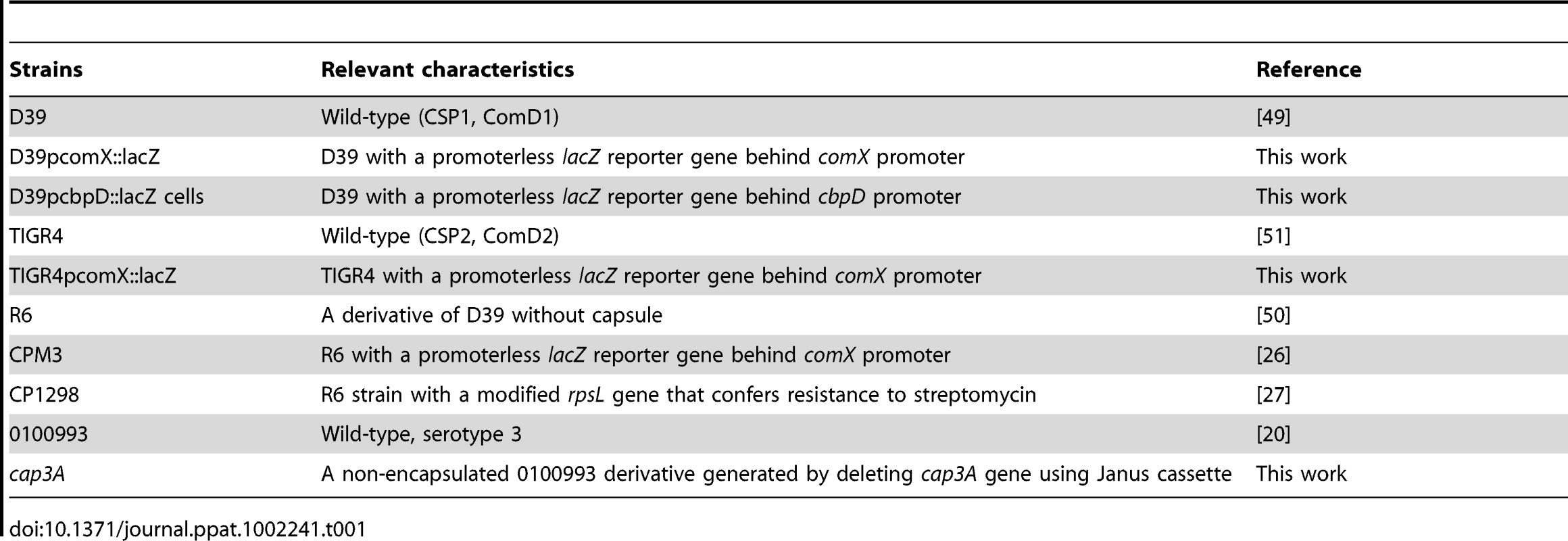
CSP1-E1A but not CSP1-R3A out-competes and inhibits CSP1-mediated comX induction in a dose and time dependent manner
As presented in Figure 2, two modified CSP peptides—CSP-E1A and CSP-R3A—are attenuated in their ability to induce the comX gene and the competence of DNA transformation. We examined the ability of these two peptides to competitively inhibit the induction of competence by CSP1. D39pcomX::lacZ cells were exposed to 100 ng of CSP1 alone or simultaneously to increasing amounts of CSP1-E1A (Fig. 3A) or CSP1-R3A (Fig. 3B). In the absence of CSP1-E1A, CSP1 induced the promoter activity of comX gene to approximately 45 Miller units of b-galactosidase. However, in the presence of CSP1-E1A, the ability of CSP1 to induce comX (Fig. 3A) was severely inhibited in a concentration dependent manner. CSP1-E1A by itself only induced low expression of comX. In contrast, even though CSP1-R3A by itself was unable to induce the expression of comX, it did not competitively inhibit CSP1-dependent comX expression (Fig. 3B). These results suggest that CSP1-E1A has the unique ability to competitively inhibit the ability of CSP1 to induce competence.
Fig. 3. Dose dependent inhibition of comX expression and DNA transformation by CSP-E1A. 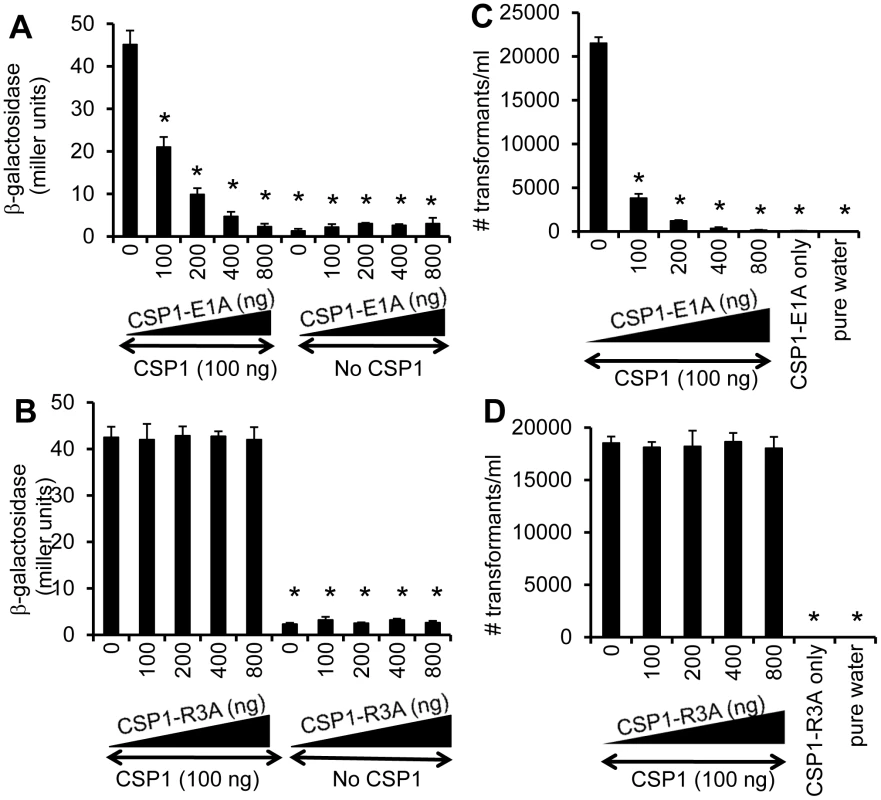
(A–B) CSP-E1A but not CSP1-R3A inhibits CSP1-mediated induction of comX in a dose dependent manner. D39pcomX::lacZ cells were exposed to 100 ng/ml of CSP1 alone or simultaneously with increasing concentrations of CSP-E1A or CSP-R3A. The activity of comX gene promoter was measured by b-galactosidase activity. (A) CSP-E1A competitively inhibits the ability of CSP1 to induce comX. (B) CSP-R3A is unable to induce comX, and does not competitively inhibit CSP1. (C–D) CSP-E1A but not CSP1-R3A outcompetes and inhibits CSP1-mediated genetic transformation in a dose dependent manner. In vitro transformation was performed by treating D39 with 100 ng/ml CSP1 alone or simultaneously with increasing concentrations of CSP1-E1A or CSP1-R3A. Purified PCR product harboring a mutated rpsL gene was used for genetic transformation. Transformants were selected on THB agar plates supplemented with 100 µg/ml streptomycin. CSP1-E1A competitively inhibited the ability of CSP1 to induce DNA transformation (C). CSP1-R3A was unable to competitively inhibit DNA transformation (D). Experiments were performed in triplicates and repeated three times. The means ± SD of one typical experiment are shown. *p<0.01 when compared against cells treated with CSP1 only. CSP-E1A but not CSP1-R3A inhibits CSP1-mediated genetic transformation
Because of their inability to induce competence and DNA transformation, we next examined whether CSP1-E1A and CSP1-R3A were able to competitively inhibit CSP1-mediated DNA transformation. In vitro transformation with the rpsL gene was performed with S. pneumoniae D39 cells exposed to 100 ng of CSP1 alone or simultaneously in the presence of increasing amounts of CSP1-E1A or CSP1-R3A. As expected, in the absence of CSP1-E1A, CSP1 induced DNA uptake and transformation at high frequency (Fig. 3C). However, the presence of increasing concentrations of CSP1-E1A drastically inhibited the transformation of D39. For example, at equal concentration of 100 ng/ml, CSP1-E1A reduced CSP1-mediated transformation by 5.6-fold. Interestingly, increasing concentration of CSP1-R3A failed to competitively inhibit the ability of CSP1 to induce DNA transformation in D39 cells (Fig. 3D). These results are consistent with the inability of CSP1-R3A to inhibit the induction of comX by CSP1 (Fig. 3B). Collectively, these data suggest that CSP1-E1A is uniquely capable of competitively inhibiting CSP1-mediated competence induction and DNA transformation.
CSP1-E1A inhibits the competence regulon in a time-dependent manner
Because CSP1-E1A is capable of competitively inhibiting CSP1-mediated competence induction in S. pneumoniae, we examined whether this inhibition occurred in a time-dependent manner. D39pcomX::lacZ cells were pre-exposed to 100 ng/ml CSP1, then chased by the addition of CSP1-E1A at indicated time intervals. Activation of the comX promoter was calculated against D39pcomX::lacZ cells exposed to CSP1 alone (100%). As shown in Figure 4, CSP1-E1A at concentration of 100 ng/ml was able to inhibit approximately 40–50% of comX induction 10 minutes after pre-induction with CSP1. The inhibition was abolished at approximately 30 minutes after preinduction with CSP1. At the concentration of 800 ng/ml, CSP1-E1A was highly effective, and able to inhibit 50% of CSP1-mediated comX induction 25 min after pre-induction by CSP1. These results suggest that under our in vitro experimental conditions, 30 min was roughly the time span required for the distribution and binding of CSP1 to ComD to prevent competitive inhibition by CSP1-E1A. In addition, these results also indicate that 30 minutes was approximately the amount of time required for establishment of full competence when S. pneumoniae D39 cells are exposed to CSP1.
Fig. 4. Time-dependent inhibition of CSP1 by CSP1-E1A. 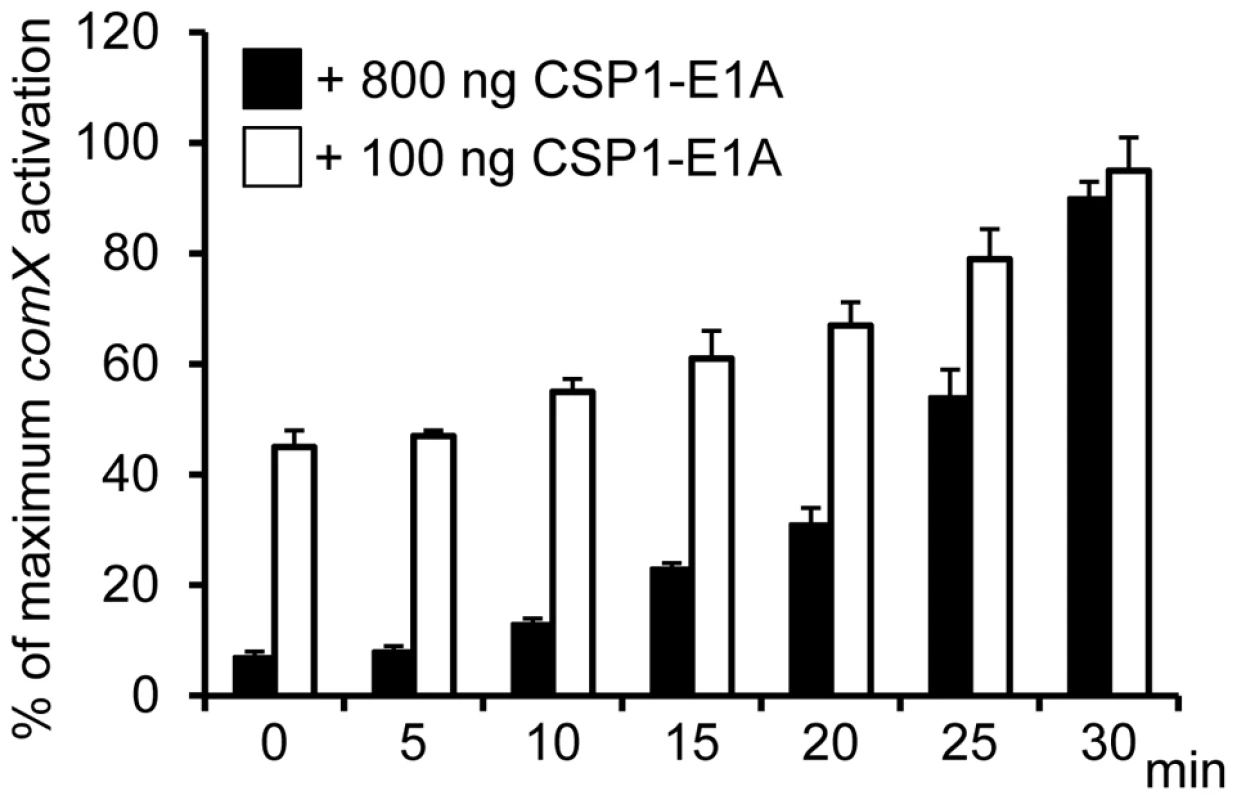
CSP1-E1A competitively inhibits CSP1 in a time-dependent manner. D39pcomX::lacZ cells (OD 600nm 0.1) were pre-exposed to 100 ng/ml CSP1. At Time 0, 5, 10, 15, 20, 25 and 30 min, CSP-E1A (100 ng or 800 ng) was added to chase CSP1. Cells were collected 30 min after each addition of CSP1-E1A for b-galactosidase assays. Activation of comX promoter was calculated against D39pcomX::lacZ cells exposed to CSP1 alone (100%). Experiments were performed in triplicates and repeated three times. The means ± SD of one typical experiment are shown. CSP1-E1A delays and inhibits the development of spontaneous competence in vitro in a concentration dependent manner
During its normal growth, S. pneumoniae cells spontaneously enter a developmental stage of competence for DNA uptake and transformation by activating the biosynthesis of CSP [27]. Because CSP1-E1A competitively inhibits the competence development in pneumococcal cells exposed to exogenously supplied CSP1, we further investigated its ability to suppress spontaneous development of competence. D39pcomX::lacZ cells grown in THB (pH 8.3) were exposed to sterile water (control) or to CSP1-E1A. The promoter activity of comX began to increase dramatically in the D39pcomX::lacZ cells incubated with sterile water at approximately 90 min (Fig. 5A). In contrast, in the D39pcomX::lacZ cells treated with 100 ng or 1 µg of CSP1-E1A, the induction of comX promoter was delayed for 30 min and 60 min, respectively (Fig. 5A). Furthermore, the maximum level comX promoter activity was only 38% in cells treated with 1 µg of CSP1-E1A when compared to those cells treated with sterile water. CSP1-E1A at the concentration of 10 µg/ml completely abolished the induction of comX promoter in D39 (Fig. 5A). Similarly, CSP1-E1A delayed and inhibited the promoter activity of a virulence gene cbpD, a late gene in the competence regulatory circuit, in a concentration dependent manner (Fig. 5B). CSP1-E1A did not interfere with the growth D39 cells (Fig. 5C). These results suggest that CSP1-E1A has the ability to delay and inhibit spontaneous competence development during normal growth in S. pneumoniae in a concentration-dependent manner.
Fig. 5. CSP1-E1A delays and inhibits the spontaneous development of competence in S. pneumoniae in a concentration-dependent manner. 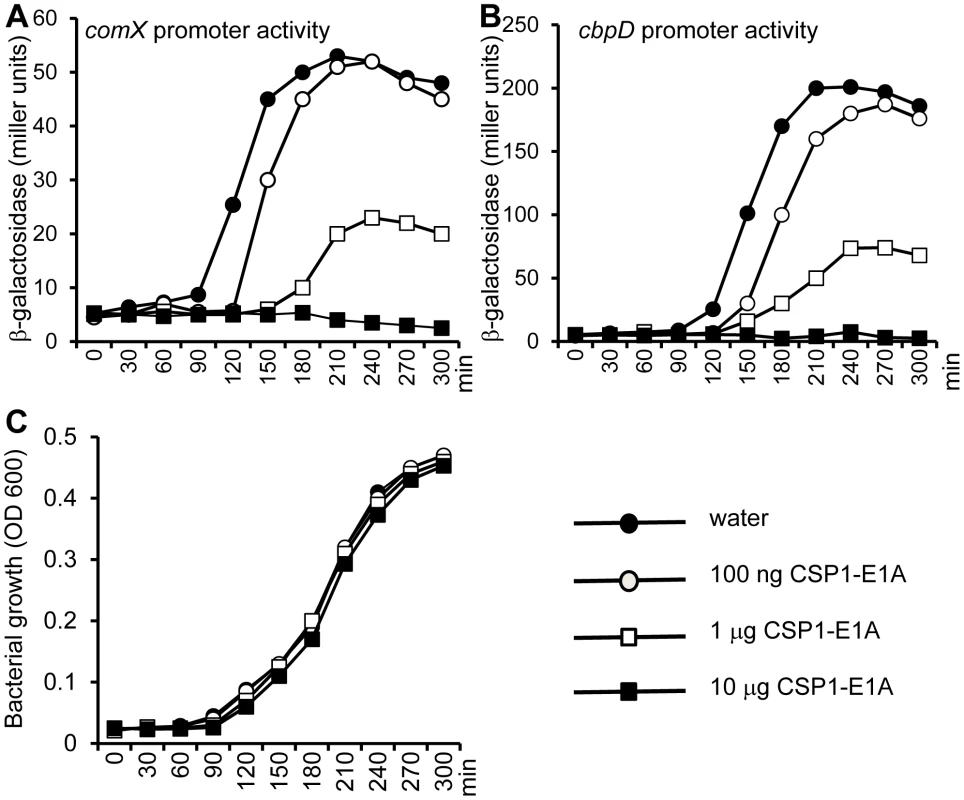
(A–B) D39pcomX::lacZ and D39pcbpD::lacZ cells grown in THB (pH 8.3) were incubated with 100 ng/ml, 1 µg/ml or 10 µg/ml of CSP1-E1A or with same volume of sterile water, and monitored for comX promoter activity (A) or for the cbpD promoter activity (B) by b-galactosidase assays at indicated time intervals. (C) CSP1-E1A does not inhibit the growth of S. pneumoniae. Similar results were obtained from three independent experiments. The data from one typical experiment are shown. CSP1-E1A attenuates the expression of S. pneumoniae virulence factor in vitro
We and others have previously shown that the competence regulon regulates both DNA transformation and virulence [20]–[24], although the mechanism behind this regulation is not fully understood. Guiral et al [23] have proposed a possible connection between competence induced cell lysis and virulence. Induction of competence triggers the expression of the murein hydrolases autolysin A (LytA) and Choline-Binding Protein D (CbpD). Together, they induce pneumococcal cell lysis and the release of virulence factors including penumolysin and cell wall component LTA into the hosts. Therefore, we examined whether CSP1-E1A could inhibit the synthesis and the release of pneumococcal murein hydrolases. S. pneumoniae D39 cells were treated with CSP1 (100 ng/ml) alone or simultaneously with increasing amounts of CSP1-E1A (0, 100, 200, 400 or 800 ng/ml), and subjected to zymogram analysis. As shown in Figures 6A–6C, The expression of CbpD and LytA decreased by 63% and 44% respectively when D39 cells were simultaneously treated with a combination of 100 ng/ml CSP1 and 200 ng/ml CSP1-E1A. The decrease is even more profound as the concentrations of CSP1-E1A was increased to 400 ng/ml or 800 g/ml, reducing the expression of LytA by 73% and 72% (Fig. 6B). Similarly, at the concentrations of 400 ng/ml or 800 g/ml, CSP1-E1A reduced the expression of CbpD by 87% and 94%, respectively (Fig. 6C). Inhibition of cpbD promoter activity (Fig. 6D) mirrored the zymogram analysis. These results suggest that CSP1-E1A is able to inhibit the expression of virulence genes regulated by competence regulon.
Fig. 6. CSP1-E1A inhibits the expression of competence regulated virulence factors LytA and CbpD in vitro. 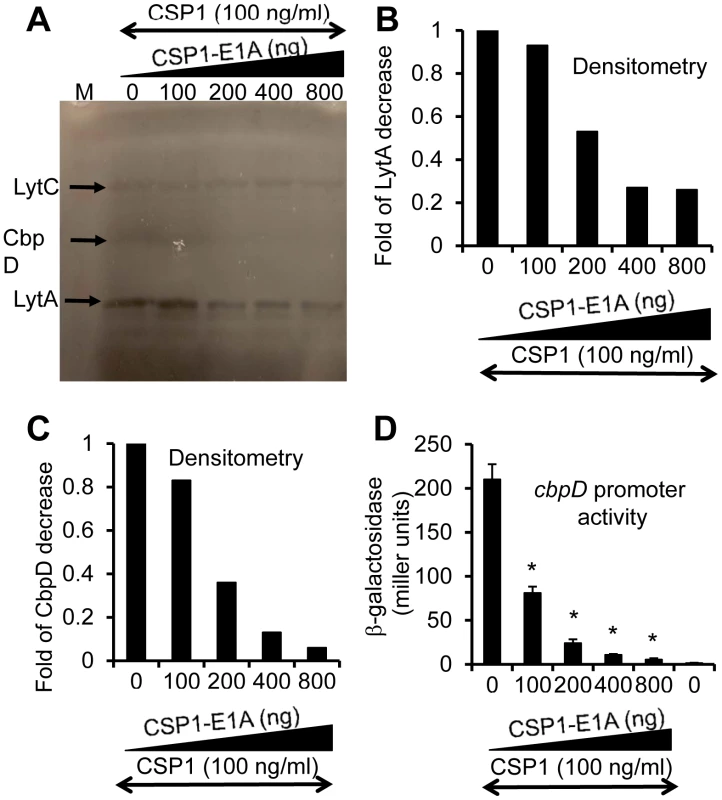
(A) CSP1-E1A attenuates the expression of LytA and CbpD. D39 cells (OD 600nm 0.1) were exposed to CSP1 (100 ng/ml) alone or simultaneously to increasing amounts of CSP1-E1A (0, 100, 200, 400 or 800 ng/ml). Cells were incubated for 30 minutes, lysed and subjected to zymogram analysis. Lytic activity was observed as bands of clear zones in the turbid gel. Experiments were repeated three times. The gel from one typical experiment is shown. (B–C) Densitometry analysis of LytA (B) and CbpD (C) from the zymogram in A. (D) CSP1-E1A inhibits the promoter activity of cbpD in a concentration dependent manner. D39pcbpD::lacZ cells were exposed to CSP1 alone, pure water alone or to a mixture of 100 ng/ml CSP1 and CSP-E1A.Cells were collected for b-galacosidase assays 30 min after exposure. CSP1-E1A attenuates the virulence of S. pneumoniae during infection of mouse lungs
The ability of CSP1-E1A to inhibit the expression of LytA and CbpD in vitro reveals the possibility of using this peptide analogue to attenuate pneumococcal infection. We examined whether CSP1-E1A could attenuate the virulence of S. pneumoniae D39 using mouse model of acute pneumonia in two independent experiments. In Experiment 1, adult CD1 mice (groups of 10) were intranasally infected with D39 alone or a mixture of D39 and 20 µg CSP1-E1A. Animals were monitored closely, and those that were moribund were considered dead. As shown in Figure 7A, inclusion of CSP1-E1A in the infection mix significantly reduced the mortality rate of infected mice from 60% (D39 alone) to 30% (D39 coinoculated with CSP1-E1A). However, the difference in kinetics of mouse survival between the two groups was not statistically significant (p = 0.21) as analyzed by the log rank (Mantel-Cox) survival analysis. Thus, we repeated the experiment with higher number of mice and increased concentration of CSP1-E1A. In Experiment 2, CD1 mice (groups of 20) were intranasally infected with D39 alone or a mixture of D39 and 100 µg CSP1-E1A. As shown in Figure 7B, increasing the CSP1-E1A concentration to 100 µg/mouse in the infection mix significantly reduced the mortality rate of infected mice from 75% (D39 alone) to 40% (D39 coinoculated with CSP1-E1A). In addition, the kinetics of death was delayed, and was statistically significant (p = 0.01) (Fig. 7B). In contrast, the survival rate and mortality kinetics were indistinguishable between mice infected with D39 alone or in mice infected with a mixture of D39 and 100 µg CSP1-R3A (Fig. 7C) (p = 0.96). These results suggest that CSP1-E1A is uniquely able to attenuate virulence mechanisms regulated by the competence regulon during S. pneumoniae-mediated lung infections.
Fig. 7. CSP1-E1A attenuates the mortality of mice infected with S. pneumoniae. 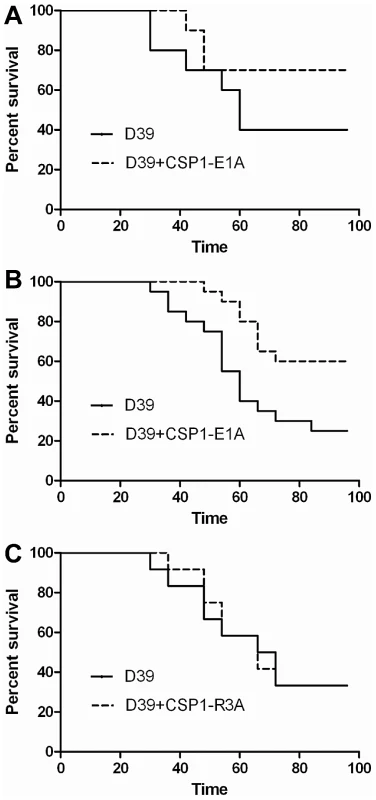
(A) Six-week old CD-1 mice (n = 10) were intranasally infected with S. pneumoniae D39 (3×106, 50 µl) alone, or with a mixture of D39-CSP-E1A (20 µg/mouse). Infected mice were monitored every 6 hr. Moribund mice were euthanized and considered dead. The difference in kinetics of mouse survival between the two groups was not statistically significant (p = 0.21) as analyzed by the log rank (Mantel-Cox) survival analysis. (B) Infection assay was repeated with CD-1 mice (n = 20) using D39 (2×106, 50 µl) alone or with a mixture of D39-CSP-E1A (100 µg/mouse). Infected mice were monitored as described in (A). The kinetics of death was significantly delayed (p = 0.01) as determined by the log rank (Mantel-Cox) survival analysis. (C) The survival kinetics was indistinguishable between mice infected with D39 alone or in mice infected with a mixture of D39 and 100 µg CSP1-R3A (p = 0.96). CSP1-E1A inhibits the horizontal gene transfer in vivo
The competence regulon plays a role in the acquisition and spread of antibiotic resistant genes in S. pneumoniae [7]–[9]. In vivo gene uptake and transformation of S. pneumoniae have been previously reported to occur at extremely low frequency [28]–[31]. As we have shown above, CSP1-E1A has the ability to competitively inhibit CSP1-mediated DNA transformation, and attenuate the development of spontaneous competence in S. pneumoniae in vitro. Here, we examined whether CSP-E1A could inhibit acquisition of the streptomycin resistance rpsL gene in vivo, using mouse models of acute pneumonia and bacteremia that we had previously described [20]. Adult CD1 mice were intranasally or intraperitoneally infected with a transformation mixture composed of D39 bacteria and rpsL donor DNA and CSP1, or a transformation mixture composed of D39 bacteria and rpsL donor DNA and CSP1 and CSP1-E1A. To ensure that no transformation occurred outside animals, each component of the transformation mixture was kept on ice. Individual transformation components were mixed on ice and immediately inoculated into the animals. Plating of the ice-cold transformation mixture before infection or at time intervals 10, 20 and 30 min after infection did not give rise to any transformants (data not shown). Intranasally-infected mice were incubated for 6 hr whereas intraperitoneally-infected mice were incubated until moribund (∼16–24 hr). As shown in Table 2, all five mice intranasally-infected with the D39-rpsL DNA-CSP1 mixture gave rise to a range of 3 to 41 streptomycin resistant transformants out of the total lung output of 106–107 D39 cells. In contrast, no streptomycin resistant D39 was recovered in the five mice infected with D39-rpsL DNA-CSP1-CSP1-E1A mixture. Similarly as shown in Table 3, all four mice intraperitoneally-infected with the D39-rpsL DNA-CSP1 mixture gave rise to a range of 273 to 517 streptomycin resistant transformants out of the total 108 D39 cells. In contrast, no streptomycin resistant D39 cells were recovered in the four mice intraperitoneally-infected with D39-rpsL DNA-CSP1-CSP1-E1A. Collectively, these results suggest that CSP1-E1A is efficient in suppressing the ability of CSP1 to activate competence regulon in vivo.
Tab. 2. CSPP1-E1A inhibits the ability of S. pneumoniae to acquire the rpsL streptomycin resistance gene in mouse lungs during acute pneumonia. 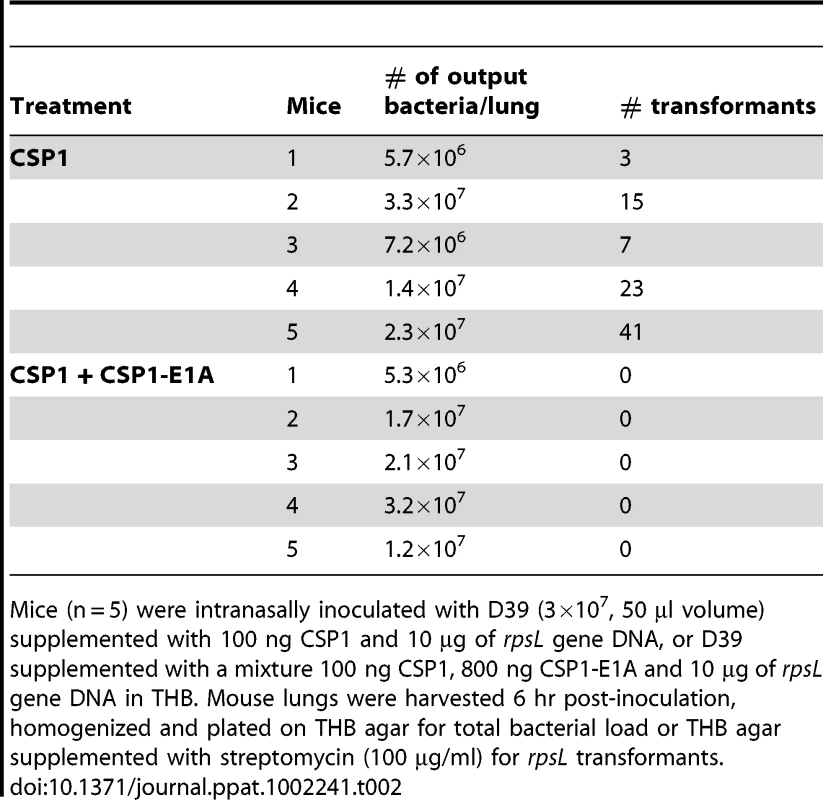
Mice (n = 5) were intranasally inoculated with D39 (3×107, 50 µl volume) supplemented with 100 ng CSP1 and 10 µg of rpsL gene DNA, or D39 supplemented with a mixture 100 ng CSP1, 800 ng CSP1-E1A and 10 µg of rpsL gene DNA in THB. Mouse lungs were harvested 6 hr post-inoculation, homogenized and plated on THB agar for total bacterial load or THB agar supplemented with streptomycin (100 µg/ml) for rpsL transformants. Tab. 3. CSPP1-E1A inhibits the ability of S. pneumoniae to acquire the rpsL streptomycin resistance gene in mouse peritoneal cavity. 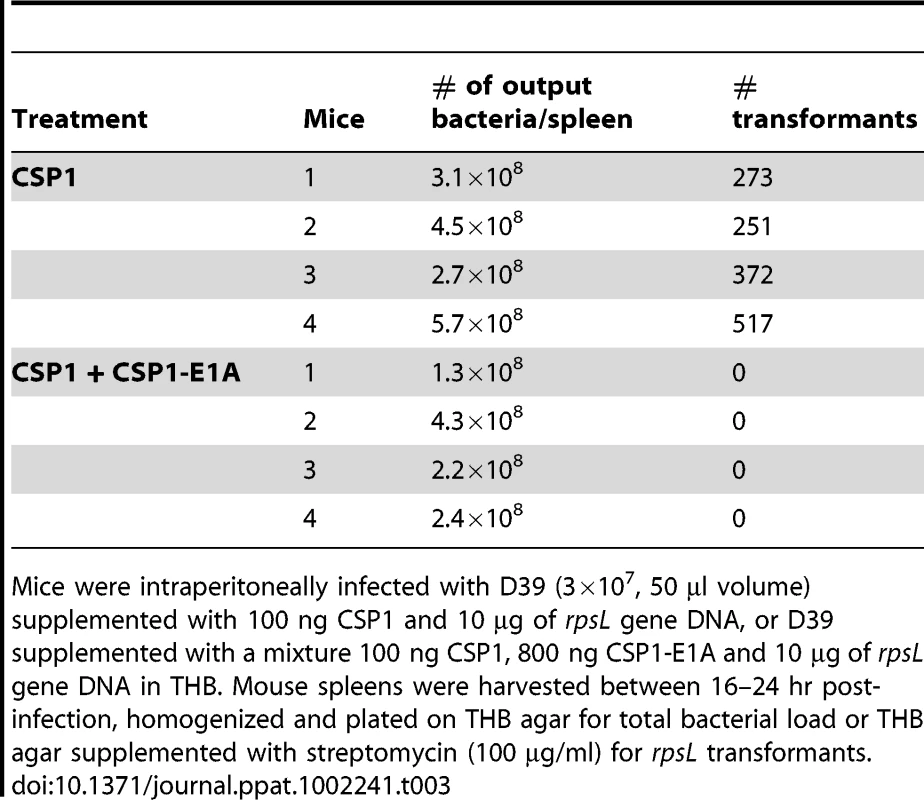
Mice were intraperitoneally infected with D39 (3×107, 50 µl volume) supplemented with 100 ng CSP1 and 10 µg of rpsL gene DNA, or D39 supplemented with a mixture 100 ng CSP1, 800 ng CSP1-E1A and 10 µg of rpsL gene DNA in THB. Mouse spleens were harvested between 16–24 hr post-infection, homogenized and plated on THB agar for total bacterial load or THB agar supplemented with streptomycin (100 µg/ml) for rpsL transformants. Spontaneous transformation of encapsulated S. pneumoniae strains including D39 occurs at extremely low frequency in vivo. Thus, CSP1 was included to increase the efficiency of DNA transformation experiments reported in Tables 2 and 3. Inclusion of CSP1 beckoned the question of whether CSP1-E1A could attenuate natural transformation events in vivo. Natural transformation experiments were performed using a capsule mutant cap3A (Table 1) derived from a serotype 3 isolate 0100993 [20]. The colonies of serotype 3 S. pneumoniae isolates are large and mucoid, and can be easily distinguished from small colonies formed by non-encapsulated rough mutants. Mice were intraperitoneally inoculated with a mixture of 3×109 cap3A cells and 100 µg of chromosomal DNA from 0100993, or with a mixture of 3×109 cap3A cells and 100 µg of 0100993 chromosomal DNA and 100 µg of CSP1-E1A. Higher amount of CSP1-E1A was used because we did not know the length of the inhibitory effect of this peptide analogue in vivo. In addition, the timing of S. pneumoniae cells entering the competent state for DNA transformation in vivo was also not clear. Transformation mixtures were assembled under ice-cold condition as described for Tables 2 and 3. Plating of transformation mixtures immediately before and after inoculation did not recover any transformants (data not shown). As shown in Table 4, transformants with wild-type capsule were recovered from four of the five mice inoculated with the mixture of cap3A bacteria and 0100993 genomic DNA. In contrast, transformants with wild-type capsule were only recovered from one out of five mice co-inoculated with cap3A bacteria and 0100993 DNA and CSP1-E1A. Furthermore, we used CSP1-R3A (Fig. 1) as a control to demonstrate the specificity of CSP1-E1A inhibition. CSP1-R3A was unable to inhibit development of natural competence in vivo (Table 4). In the group treated with a mixture of 3×109 cap3A cells and 100 µg of 0100993 chromosomal DNA and 100 µg of CSP1-R3A, transformants were isolated from all the 5 mice, indicating high concentration of CSP1-R3A was unable to inhibit natural DNA transformation in vivo. These results suggest that CSP1-E1A has the unique ability to reduce the incidence of natural DNA transformation process in vivo.
Tab. 4. CSPP1-E1A attenuates the ability of cap3A mutant S. pneumoniae to acquire capsule gene during natural transformation in mouse peritoneal cavity. 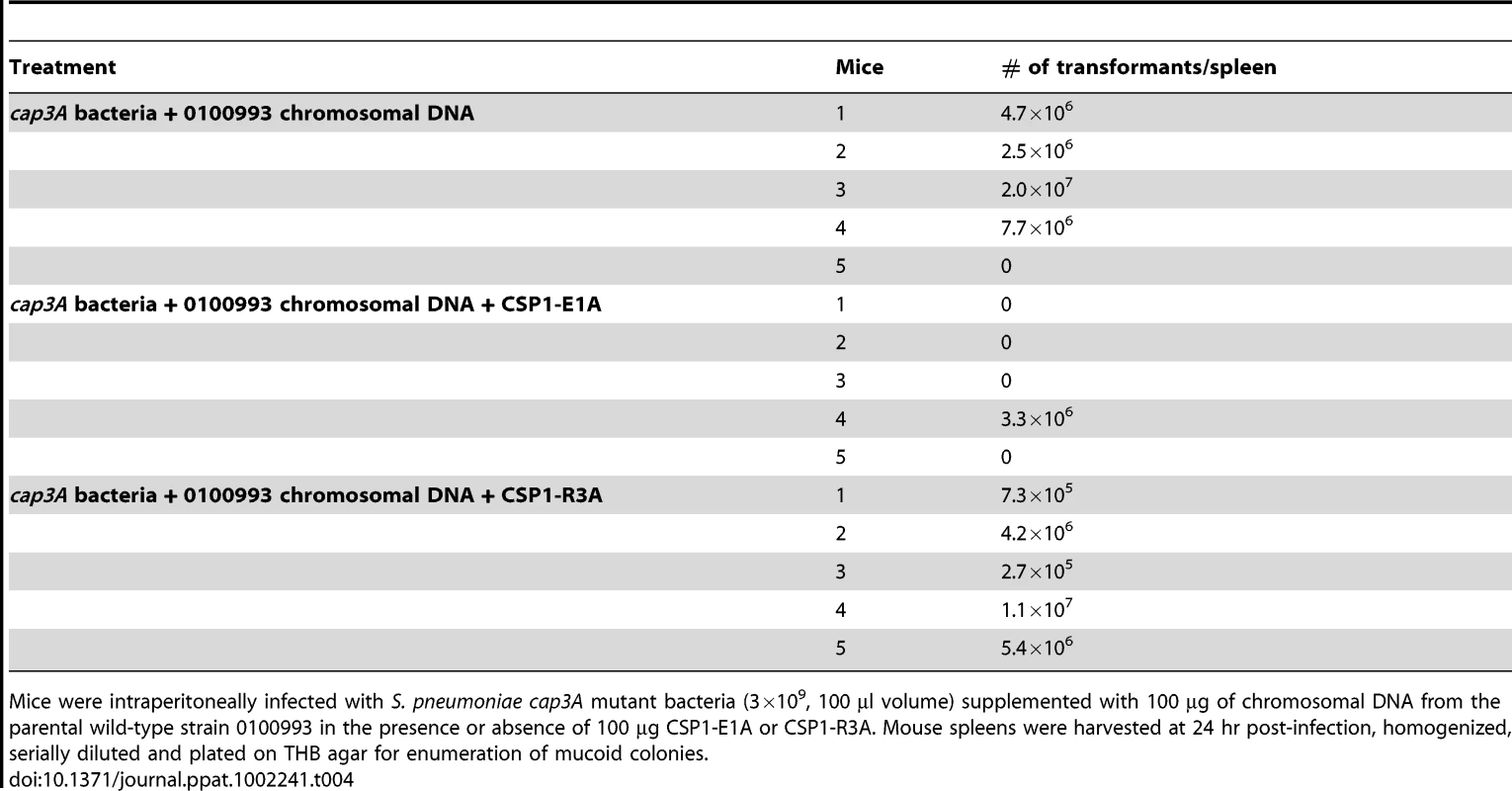
Mice were intraperitoneally infected with S. pneumoniae cap3A mutant bacteria (3×109, 100 µl volume) supplemented with 100 µg of chromosomal DNA from the parental wild-type strain 0100993 in the presence or absence of 100 µg CSP1-E1A or CSP1-R3A. Mouse spleens were harvested at 24 hr post-infection, homogenized, serially diluted and plated on THB agar for enumeration of mucoid colonies. CSP1-E1A is not bacteriostatic against S. pnuemoniae
A previous study has shown that CSP2 has bacteriostatic activity against S. pneumoniae [32]. To rule out non-specific effects due to a much higher concentration of CSP1-E1A used in the virulence attenuation (Fig. 7B) and in vivo natural transformation (Table 4) studies, we examined whether CSP1-E1A was bacteriostatic against S. pneumoniae. As shown in Figure S1A, CSP1 at concentrations of 20 µg or 100 µg caused a slight delay in the growth of D39 cells. In contrast, same concentrations of CSP1-E1A or CSP1-R3A did not slow the growth of D39 cells (Fig. S1B–S1C). These results suggest that high concentrations of CSP1-E1A is not bacteriostatic, and that attenuation of virulence and DNA transformation by CSP1-E1A is due to inhibition of competence regulon induction.
A CSP2 analogue CSP2-E1A effectively inhibits the competence development and transformation in ComD2 strain TIGR4
Because CSP1-E1A is an efficient inhibitor of competence induced by CSP1, we hypothesize that a synthetic CSP2 analogue with the substitution of first residue (glutamate) with alainine (CSP2-E1A) will competitively inhibit CSP2 (Fig. 1). We examined the ability of CSP2-E1A to inhibit the induction of competence by CSP2 in the compatible ComD2 strain TIGR4 (Table 1). CSP2-E1A was barely able to induce the promoter activity of comX (Fig. 8A), and its ability to induce DNA transformation was reduced by approximately 3 logs when compared to CSP2 (Fig. 8B). Importantly, CSP2-E1A competitively inhibited the ability of CSP2 to induce comX expression in the TIGR4pcomX::lacZ cells in a concentration-dependent manner (Fig. 8C). Similarly, CSP2-E1A inhibited the ability of CSP2 to induce DNA transformation of TIGR4 in a concentration dependent manner. At equal concentration of 100 ng/ml, CSP2-E1A reduced the number of transformants generated by CSP2 by 68% (Fig. 8D). At higher concentrations of 400 and 800 ng/ml, CSP2-E1A almost completely abolished CSP2-mediated DNA transformation. These results suggest that CSP2-E1A is a strong competitive inhibitor of comX expression and DNA transformation in ComD2 strain TIGR4.
Fig. 8. CSP2-E1A inhibits competence development and transformation in ComD2 strain TIGR4. 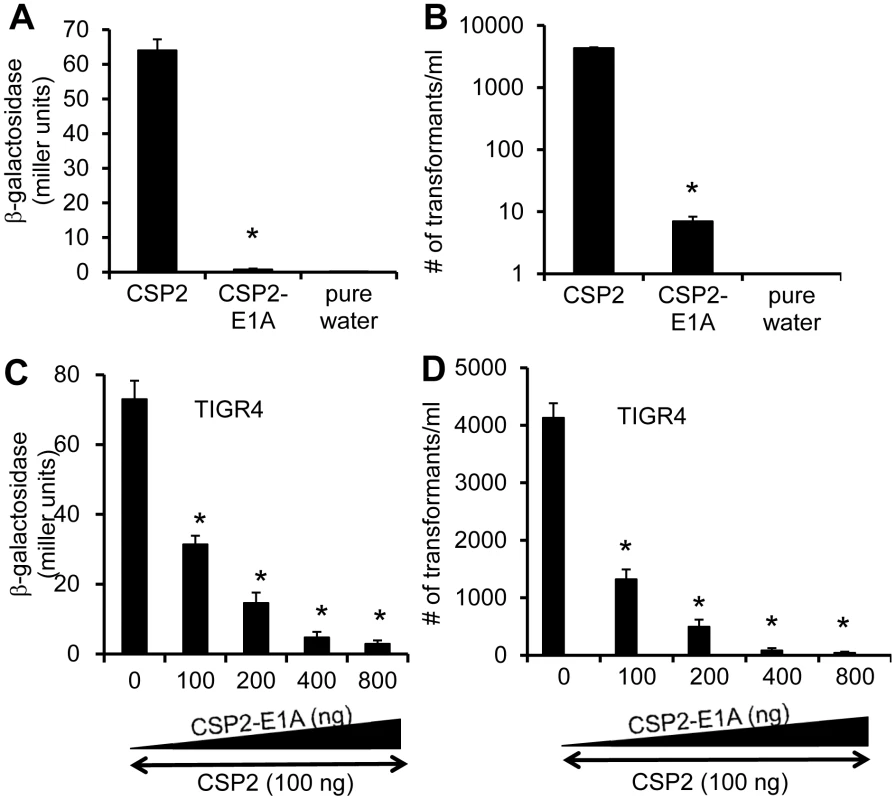
(A–B) CSP2-E1A was drastically reduced in its ability to induce the promoter activity of comX (A) and DNA transformation in TIGR4 (B). Experiments were performed in triplicates and repeated three times. The means ± SD of one typical experiment are shown. *p<0.01 when compared against TIGR4pcomX::lacZ cells (A) or TIGR4 cells (B) treated with CSP2 only. (C) CSP-E2A out-competes CSP2 and inhibits activation of the comX promoter activity. TIGR4pcomX::lacZ cells (OD 600nm 0.1) were incubated with 100 ng/ml CSP2 alone or simultaneously with increasing amounts of CSP2-E1A. The activity of comX promoter was measured by b-galactosidase assays. (D) CSP-E2A inhibits CSP2-mediated DNA transformation of TIGR4. In vitro transformation was performed by treating TIGR4 cells with 100 ng/ml CSP2 alone or simultaneously with increasing amount of CSP2-E1A. Purified PCR product of a mutated rpsL gene was used for genetic transformation. Transformants were selected on THB agar supplemented with 100 µg/ml streptomycin. Experiments in were performed in triplicates and repeated three times. The means ± SD of one typical experiment are shown. *p<0.01 when compared against TIGR4pcomX::lacZ cells (A) or TIGR4 cells (B) treated with CSP2 only. CSP1-E1A and CSP2-E1A cross inhibit the competence in S. penumoniae strains with incompatible ComD subtypes
A previous study has shown that individual CSP peptide is capable of inducing the competence of DNA transformation in S. pneumoniae strains expressing an incompatible ComD subtype, albeit at lower efficiency [19]. For example, CSP1 is able to induce the competence at lower efficiency in TIGR4, a pneumococcal strain with an incompatible ComD2 subtype. Because the first amino acid (glutamate) at the N-terminus is conserved across all six CSP subtypes, this raises the possibility that our CSP analogues CSP1-E1A and CSP2-E1A can be broad spectrum in their ability to inhibit the development of competence, horizontal gene transfers and virulence, regardless of S. pneumoniae serotypes or ComD subtypes. To test this hypothesis, we examined the ability of CSP1-E1A and CSP2-E1A (Table 1) to cross inhibit the induction of comX and DNA transformation in ComD2 strain TIGR4 or ComD1 strain D39, respectively. As shown in Figure 9A, CSP1-E1A inhibited the induction of comX and DNA transformation by CSP2 in TIGR4 by a concentration dependent manner, though at a lesser efficiency. For example, CSP1-E1A at 800 ng/ml was able to inhibit approximately 50% of comX promoter activity in the presence of 100 ng/ml CSP2. Similarly, CSP1-E1A (800 ng/ml) decreased the CSP2-induced DNA transformation frequency by 67% (Fig. 9B).
Fig. 9. Cross inhibition of competence by CSP1-E1A and CSP2-E1A in S. pneumoniae strains with incompatible ComD subtypes. 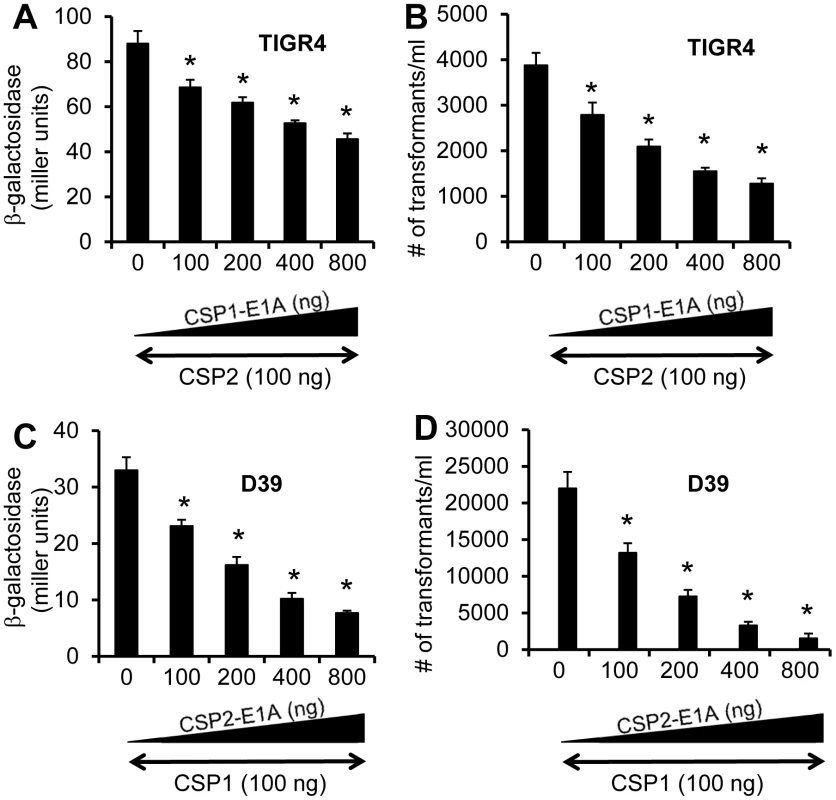
(A-B) TIGR4 (ComD2 subtype) or TIGR4pcomX-lacZ cells (OD 600nm 0.1) were incubated with CSP2 alone or simultaneously with increasing amounts of CSP1-E1A (100–800 ng/ml). (C-D) D39 (ComD1 subtype) or D39pcomX-lacZ cells were incubated with the mixture of 100 ng/ml CSP1 alone or simultaneously with increasing amounts of CSP2-E1A (100–800 ng/ml). Pneumococcal cells were assayed for ß-galactosidase activity and transformation frequency 60 minutes after exposure. (A–B) Cross inhibition of competence in ComD2 strain TIGR4 by CSP1-E1A as measured by the induction of comX promoter activity (A) and transformation frequency (B). (C–D) Cross inhibition of competence in ComD1 strain D39 by CSP2-E1A as measured by the induction of comX promoter activity (C) and transformation frequency (D). Experiments were performed in triplicates and repeated three times. The means ± SD of one typical experiment are shown. *p<0.05 when compared against TIGR4 cells or TIGR4pcomX-lacZ cells treated with CSP2 alone, or D39 cells or D39pcomX-lacZ cells treated with CSP1 alone. Finally, we examined the ability of CSP2-E1A to inhibit the induction of comX and DNA transformation by CSP1 in the ComD1 strain D39. As shown in Figure 9C, CSP2-E1A inhibited CSP1-mediated comX induction and DNA transformation in D39 by a concentration dependent manner. For example, CSP2-E1A at 800 ng/ml was able to inhibit approximately 77% of comX induction and 90% of DNA transformation in the presence of 100 ng/ml CSP1 (Fig. 9D). Collectively, these results suggest that CSP1-E1A and CSP2-E1A have the potential of inhibiting the development of competence, horizontal gene transfer and virulence, regardless of pneumococcal serotypes and CSP/ComD subtypes.
Discussion
The competence regulon is known to regulate DNA transformation and virulence in S. pneumoniae [7]–[9], [20]–[24]. In this study, we explored the use of synthetic analogues of CSPs to competitively inhibit: (i) the development of competence, (ii) horizontal gene transfer, and (iii) virulence, under both in vitro and in vivo experimental conditions. Amino acid substitution or deletion of conserved residues in C-terminus of CSP1 did not alter their ability to induce competence and DNA transformation. In contrast, amino acid substitutions on the first (CSP1-E1A) and third residue (CSP1-R3A) on the N-terminus impair the ability of these CSP analogues to induce competence. Our findings are consistent with the previously thesis work by Coomaraswamy [33], which has been summarized by Haverstein and Morrison [34]. By alanine scanning, Coomaraswamy demonstrates that both the N-terminus and the central region are more important than the C-terminus of CSP1 for the induction of competence regulon. One discrepancy of our finding from that of Coomaraswamy is on the requirement of 2nd amino acid residue on the N-terminus. Coomaraswamy has shown that substituting the methionine with an alanine reduces the ability of the CSP1-M2A to induce competence by 96%. In contrast, our analysis suggests that the methionine residue is dispensable for competence induction. One possible explanation for the discrepancy is that our comX induction and DNA transformation experiments were performed with 100 ng/ml CSP1-M2A, approximately 10-fold higher than the amounts used by Coomaraswamy. Thus, despite its lower activity, excess amounts of CSP1-M2A might have masked our ability to see reduced efficiency of this analogue to induce competence.
Of the five CSP1 analogues examined, only CSP1-E1A is capable of competitively inhibiting the induction of competence and DNA transformation by CSP1. CSP1-E1A is able to effectively compete with CSP1 even after 25 min pre-exposure of S. pneumoniae to the latter. Furthermore, CSP1-E1A is able to inhibit the development of spontaneous competence in S. pneumoniae in a concentration dependent manner. Importantly, CSP1-E1A is able to attenuate the expression of competence-regulated pneumococcal virulence factors LytA and CbpD in vitro, and reduced the mortality of mice in a pneumonia model of lung infection by S. pneumoniae. In addition, we provide evidence that CSP1-E1A is able to inhibit the horizontal gene transfer of the streptomycin resistance rpsL and the capsule cap3A genes in vivo. Finally, we demonstrate a broader applicability of the peptide inhibitor strategy. CSP1-E1A and CSP2-E1A are able to cross inhibit the induction of competence and DNA transformation in pneumococcal strains with incompatible ComD subtypes. For example, CSP1-E1A is able to reduce the CSP2-mediated competence and DNA transformation in ComD2 strain TIGR4. Similarly, CSP2-E1A is able to cross inhibit the CSP1-mediated competence and DNA transformation in ComD1 strain D39.
Among the five CSP1 analogues, we found that only CSP1-E1A inhibits CSP1-mediated competence in time and concentration-dependent manners. These results suggest that glutamate, the first amino acid residue of CSPs, is only required for the activity of CSPs. Substituting the glutamate with alanine disrupts the ability of the analogue to activate ComD1, but does not interfere with its binding affinity to ComD1. Competitive inhibition of CSP1 by CSP1-E1A suggests that this analogue, which barely activates competence, can bind to the receptor kinase ComD1, and outcompete CSP1. In contrast, another CSP1 analogue, CSP1-R3A, not only loses the ability to induce competence, but also could not out compete CSP1 even when added in over-saturating concentrations. This observation indicates that CSP1-R3A has lost its ability to bind and activate ComD1. The importance of the first glutamate residue has been confirmed by our data demonstrating that the synthetic analogue CSP2-E1A also efficiently inhibits the CSP2-mediated DNA transformation in the compatible ComD2 strain TIGR4.
Several studies have revealed that the competence regulon of S. pneumoniae is important for virulence in mammalian hosts [20]–[24]. For example, we have previously shown that the loss-of-function mutation in the comB gene, which encodes an accessory protein of the ComA ABC transporter for the export of CSP, is attenuated in a mouse model of bacteremia infection [20]. Mutation of CSP receptor gene comD severely attenuates virulence in both mouse models of pneumonia and bacteremia infection [20]. Also, microarray analysis has showed that some virulence and stress responsive genes are upregulated during development of competence, suggesting that genes that are important for the fitness of S. pneumoniae are also regulated by competence CSP-ComDE cascade [14]. Thus, suppression of the CSP-ComDE cascade or downstream genes dependent on this signaling pathway may attenuate the virulence of this pathogen. Our results, which show that CSP1-E1A delays the kinetics and the rate of mouse mortality, demonstrate that this peptide has the potential to inhibit the virulence of S. pneumoniae by disrupting the CSP-ComDE - dependent competence regulon. Our findings are in agreement with previous reports by Oggioni et al, which show that administration of CSP in a mouse model of pneumonia infection increases virulence of pneumococci [35], suggesting that competence regulon is important for lung infection. In contrast, the same authors also reported that intravenous (IV) injection of CSP reduces virulence in an IV model of mouse bacteremia [32]. Based on this finding, one can surmise that administration of CSP1-E1A may have the unintended consequence of exacerbating virulence in IV model of mouse bacteremia. However, one unresolved issue with the results of the IV model of sepsis [32] is that it is in disagreement with the findings by Lau et al [20], which showed the requirement of competence regulon during pneumococcal-mediated sepsis. The discrepansy can be explained because the sepsis studies by Lau et al was performed by intraperitoneal model of infection whereas the studies by Oggionu et al was performed by IV model of infection. Neverthelss, collectively, these studies indicate the important contribution of the pneumococcal competence regulon to virulence during mouse model of pneumonia infection, and offer an tentalizing clue that the competence system as interesting drug target to combat pulmonary pneumonia. In contrast, inhibition of competence regulon by systemic delivery of CSP1-E1A in an IV model of bacteremia should be avoided until further clarification.
Within the last few decades, resistance of S. pneumoniae to β-lactams, macrolides, and other antibiotic classes has escalated dramatically throughout the world [4]–[6], [36]–[39]. While conjugative elements appear to be the most important causes for the spread of antibiotic resistance genes in S. pneumoniae [40]–[43], horizontal gene transfer of penicillin binding protein genes has occurred in clinical isolates of S. pneumoniae [44]. Importantly, it has been shown that under in vitro conditions, antibiotic stress induces genetic transformability in S. pneumoniae [45]. This suggests that under certain in vivo circumstances, instead of being killed, S. pneumoniae actively exploits the opportunity of antibiotic stress to acquire exogenous genes. These authors also reported that unlike wild type S. pneumoniae, antibiotic stress-mediated competence could not be induced in a comA mutant that lacks the ABC transporter needed to export CSP, suggesting that this process is CSP-dependent. Because antibiotic resistance is a serious clinical concern, the development and use of non-antibiotic based therapy to reduce horizontal gene transfer is urgently needed. Our studies suggest that CSP1-E1A and CSP2-E1A competitively inhibit CSP from binding to ComD, leading to suppression of horizontal gene transfer. The use of CSP1-E1A and CSP2-E1A, which does not inhibit the bacterial growth, is predicted to pose less selection pressure toward the survival of S. pneumoniae. This may alleviate the emergence of resistance in pneumococcus. More importantly, CSP1-E1A and CSP2-E1A have the ability to cross inhibit the induction of the competence regulon in S. pneumoniae strains with incompatible ComD subtypes. Thus, it may be beneficial to generate analogues of CSPs as drugs to reduce horizontal transfer of antibiotic resistance and virulence genes, and to attenuate virulence during infection by S. pneumoniae irrespective of their serotypes. In the same context, chemical inhibitors of that interfere with CSP functions may be useful as well.
In previously published studies, some of the in vivo transformation experiments involved infections were allowed to run their course, and success or failure of gene transfers were expressed by the number of deceased animals harboring encapsulated or antibiotic resistant transformants [28]–[31]. However, at these late stages of infection, most transformants are expected to have over-proliferated, and it may be difficult to assess the actual transformation frequency. Other transformation studies lasted for eight hr [46], [47]. In this study, we used both approaches. We assessed the infected lungs for streptomycin resistant transformants 6 hr post-infection in an effort to estimate the actual transformation frequency. Our results suggest that, despite the provision of CSP1 and abundant amounts of DNA, in vivo transformation events for the encapsulated wild-type S. pneumoniae strain D39 in mouse lungs remain low, in the range of 10−6 to 10−7 under our experimentation conditions. In comparison, we also examined the in vivo transformation within intraperitoneally-infected mice in which the infection was allowed to take its full course (∼ 24 hr), similar to previously published studies [28]–[31]. The percentage of transformants in D39 increases to the range of 10−4 to 10−5. The different rate of transformation may suggest that the environment in the lung is more complex and less conducive to DNA uptake and transformation than the intraperitoneal space. Alternatively, longer period of infection may have allowed more recipient D39 cells to be transformed.
Importantly, we show that CSP1-E1A is also able to attenuate natural transformation in the absence of exogenously provided CSP1. The capsule-deficient cap3A mutant derived from the serotype 3 strain 0100993 was used as recipient. It is well known that natural transformation frequency of capsule-deficient S. pneumoniae mutants is much higher than their respective encapsulated parental wild-type strains. Furthermore, recipients that have acquired the wild-type capsule gene will be more resistant to phagocytosis by host leukocytes, allowing a better possibility for the transformants to proliferate inside the host. Because of the higher transformation frequency was anticipated for the rough cap3A mutants, larger amounts of CSP1-E1A was used to attenuate natural transformation to ensure that the inhibition effect of CSP1-E1A could outlast and suppress spontaneous development of competence of cap3A cells in the peritoneal cavity. In contrast, the presence of capsule in D39 cells reduces the spontaneous transformation frequency to a very low level. Not surprisingly, 800 ng of CSP1-E1A was adequate to completely abolish transformation of D39 cells in vivo.
In summary, we have demonstrated the efficacy of using competitive inhibitor of CSPs to attenuate virulence and to inhibit horizontal gene transfer in S. pneumoniae. Because competence peptide-mediated quorum sensing systems are conserved in many pathogenic species of Gram-positive bacteria [48]–[52], the strategy of using peptide analogue may be applicable to reduce the incidence of horizontal gene transfer and acquisition of antibiotic resistance genes, and to also treat lung infections mediated by this class of pathogens. In addition, competence systems in other streptococcal species, including S. mitis, are very similar to that of S. pneumoniae, lending to the idea that the former could be transformed by pneumococcal DNA [53], [54]. S. mitis is generally considered as an avirulent species, but it has the potential to acquire virulence determinants from S. pneumoniae to transform itself into a pathogen. In this scenario, CSP1-E1A or CSP2-E1A may be applicable to reduce the emergence of the new pathogen.
Materials and Methods
Chemicals
All chemicals, except where noted, were purchased from Sigma Chemical Co. (St. Lois, MO).
Bacterial strains and growth conditions
S. pneumoniae strains D39 [55], R6 [56] and TIGR4 [57] were generous gifts from Dr. David Briles (University of Alabama-Birmingham). Strains D39pcomX::lacZ and TIGR4pcomX::lacZ, both of which carry an insertion of the lacZ gene under the control of the comX promoter, were generated by transforming D39 and TIGR4 with the genomic DNA from CPM3 [26]. Strain D39pcbpD::lacZ was generated using previously described primers [58]. Briefly, a 500 bp fragment within cbpD gene was amplified and cloned into lacZ reporter plasmid pEVP3 [59]. The recombinant plasmid was transformed into D39 to generate D39pcbpD::lacZ. The capsule-deficient cap3A mutant was generated by deleting cap3A gene from the serotype 3 clinical isolate 0100993 [20] using the Janus cassette [25]. Aliquots of bacteria were stored at −70°C in THB containing 25% glycerol. For routine experiments, bacteria from frozen stocks were streaked onto THB agar containing 5% defibrinated horse blood and incubated for 12–24 h at 37°C with 5% CO2. Fresh colonies were scraped and transferred to THB and grew to desired density as measured by a spectrophotometer at OD 600nm.
Synthetic CSPs
CSP1, CSP2 and their analogues (≥95% purity) were synthesized by Elim Biopharm (Hayward, CA).
Activation assays of comX and cbpD promoters
The ability of synthetic CSPs to activate the promoters of comX gene was compared in D39pcomX::lacZ or TIGR4pcomX::lacZ bacteria grown in THB (pH 6.8) until OD 600nm of 0.1, washed and resuspended in THB (pH 8.3). Induction of cbpD promoter was monitored using the D39pcbpD::lacZ cells grown under the same conditions. CSP1 or CSP2 and their analogues were added at indicated concentrations to the culture and incubated at 37°C for 30 min. ß-galactosidase activity was measured according to previously published protocols [19], and expressed as Miller units.
In vitro S. pneumoniae transformation assay
In vitro transformation experiments were performed as we had previously described [20]. Briefly, S. pneumoniae cells were grown to early log phase (OD 600nm 0.1) in THB (pH 6.8), washed and resuspended in THB (pH 8.3) containing rpsL DNA (1 µg). CSP1, CSP2 and their analogues were added alone or in combination at desirable concentrations. The rpsL gene, which confers resistance to streptomycin, and its flanking regions (combined length 1633 bp), were amplified from pneumococcal strain CP1296 [27] using the following primers: rpsL upper 5′-GGGCTAGTAGAAGTAGTTGG-3′; rpsL lower: 5′-CGGAAGTGTGCGAATGCACG-3′). The transformation mix was then incubated at 37°C with 5% CO2 for 1 hr. Transformants were selected on THB agar supplemented with 100 µg/mL streptomycin.
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Illinois at Urbana-Champaign (Protocol Number: 10144).
Mouse acute pneumonia infection assays
Mice used in this study were housed in positively ventilated microisolator cages with automatic recirculating water, located in a room with laminar, high efficiency particle accumulation–filtered air. The animals received autoclaved food, water, and bedding. Six-week old adult male CD-1 mice (Charles River, Boston) (groups of 10) were anesthetized with isoflurane and intranasally administered 3×106 CFU of D39 bacteria or D39-20 µg CSP1-E1A mixture in 50 µl PBS. In a second experiment, CD-1 mice (groups of 20) were exposed to 2×106 CFU of D39 bacteria or D39-100 µg CSP1-E1A mixture in 50 µl PBS. The Infected mice were monitored every 6 hr for 96 hr. Moribund animals that displayed rough hair coat, hunched posture, distended abdomen, lethargy or inability to eat or drink were euthanized.
In vivo horizontal gene transfer studies
The rpsL donor DNA was amplified from pneumococcal strain CP1296 [25]. S. pneumoniae strain D39 cells were used as recipients. D39 cells were grown to OD 600nm 0.1 in THB (pH 6.8), washed and resuspended in fresh THB (pH 8.3). Adult CD-1 mice were intranasally or intrapertoneally infected with D39 (3×107, 50 µl volume) supplemented with a mixture of 100 ng CSP1 and 10 µg of rpsL gene DNA, or D39 supplemented with a mixture 100 ng CSP1, 800 ng CSP1-E1A, and 10 µg of rpsL gene DNA in THB. For natural transformation studies, a capsule-deficient cap3A mutant (Table 1) derived from a clinical serotype 3 strain 0100993 (20) was used as recipient. Natural transformation were performed intraperitoneally in mice with 2×109 cap3A cells and 100 µg of chromosomal DNA from 0100993, with or without 100 µg of CSP1-E1A. No CSP1 peptide was used. For all transformation assays, individual reagents were kept separately on ice, mixed and immediately inoculated into animals to prevent the induction of competence prior to injection. Transformation event did not occur before or after the experiments because no transformants was recovered from plating the transformation mix before or after the experiments (data not shown). For intranasally infected mice, lungs were harvested 6hr post-infection, homogenized and plated on THB agar (total output) or THB agar supplemented with 100 µg/ml streptomycin for transformants. Intraperitoneally infected mice were monitored until they were moribund (∼16–24 hr) at which time the animals were euthanized, and the spleens were harvested for bacterial enumeration. The number of total bacteria and streptomycin resistant transformants in spleens were enumerated as described above.
S. pnuemoniae growth inhibition by CSP1 and derivatives
D39 cells were collected at early log phase (OD 600nm 0.1). Cells (2×106 cfu) were resuspended in 50 µl of sterile PBS and incubated with 20 µg or 100 µg of CSP1, CSP1-E1A or CSP1-R3A or CSP1, and incubated on ice for 10 min. THB (500 µl) was added to the cells/peptide mixtures, and incubated at 37°C in a CO2 incubator. Bacterial growth was monitored at OD 600nm using a spectrophotometer for the indicated time intervals.
Zymogram analysis of murein hydrolases
D39 cells were grown in THB (pH 6.8) to OD 600nm of 0.1. Cells were washed and resuspended in fresh THB (pH 8.3). Cells were exposed to CSP1 (100 ng/ml) alone or to a combination of CSP1 and increasing amounts of CSP1-E1A (0, 100, 200, 400 or 800 ng/ml). After 30 minutes of incubation, cells were lysed by mixing with 2X SDS loading buffer, and subjected to zymogram analysis as previously published [60]. Lytic activity was observed as bands of clear zones in the turbid gel.
Statistical analyses
Statistical analyses of in vitro experiments were performed using the Student's t-test and one-way analyses of variance (ANOVA). Statistical significance of survival studies was performed using the log-rank (Mantel-Cox) test in GraphPad Prism statistical software package. A significant difference was considered to be p<0.05.
Accession numbers
gene = “comC1”, locus_tag = “SPD_2065”, db_xref = “GeneID:4441955”;
gene = “comD”, locus_tag = “SPD_2064”, db_xref = “GeneID:4442986”;
gene = “comE”, locus_tag = “SPD_2063”, db_xref = “GeneID:4441307”;
gene = “comX1”, locus_tag = “SPD_0014”, db_xref = “GeneID:4441078”;
gene = “comX2”, locus_tag = “SPD_1818”, db_xref = “GeneID:4442416”;
gene = “cbpD”, locus_tag = “SPD_2028”, db_xref = “GeneID:4442443”;
gene = “cap3A”, GenBank: Z47210.1.
Supporting Information
Zdroje
1. WeiserJN 2010 The pneumococcus: why a commensal misbehaves. J Mol Med 88 97 102
2. MitchellAMMitchellTJ 2010 Streptococcus pneumoniae: virulence factors and variation. Clin Microbiol Infect 16 411 418
3. KadiogluAWeiserJNPatonJCAndrewPW 2008 The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol 6 288 301
4. CokerTRChanLSNewberrySJLimbosMASuttorpMJ 2010 Diagnosis, Microbial Epidemiology, and Antibiotic Treatment of Acute Otitis Media in Children: A Systematic Review. JAMA 304 2161 2169
5. LynchJP3rdZhanelGG 2010 Streptococcus pneumoniae: epidemiology and risk factors, evolution of antimicrobial resistance, and impact of vaccines. Curr Opin Pulm Med 16 217 225
6. LiñaresJArdanuyCPallaresRFenollA 2010 Changes in antimicrobial resistance, serotypes and genotypes in Streptococcus pneumoniae over a 30-year period. Clin Microbiol Infect 16 402 410
7. JohnsborgOHåvarsteinLS 2009 Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol Rev 33 627 642
8. ClaverysJPMartinBPolardP 2009 The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol Rev 33 643 656
9. MorrisonDALeeMS 2000 Regulation of competence for genetic transformation in Streptococcus pneumoniae: a link between quorum sensing and DNA processing genes. Res Microbiol 151 445 451
10. HavarsteinLSCoomaraswamyGMorrisonDA 1995 An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA 92 11140 11144
11. PestovaEVHåvarsteinLSMorrisonDA 1996 Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol 21 853 862
12. HåvarsteinLSGaustadPNesIFMorrisonDA 1996 Identification of the streptococcal competence-pheromone receptor. Mol Microbiol 21 863 869
13. MartinBGranadelCCampoNHénardVPrudhommeMClaverysJP 2010 Expression and maintenance of ComD-ComE, the two-component signal-transduction system that controls competence of Streptococcus pneumoniae. Mol Microbiol 75 1513 1528
14. PetersonSSungCKClineRDesaiBVSnesrudE 2004 Identification of competence pheromone responsive genes in Streptococcus pneumoniae. Mol Microbiol 51 1051 1070
15. LuoPLiHMorrisonDA 2003 ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol Microbiol 50 623 633
16. PozziGMasalaLLannelliFManganelliRHåvarsteinLS 1996 Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J Bacteriol 178 6087 6090
17. WhatmoreAMBarcusVADowsonCG 1999 Genetic diversity of the streptococcal competence (com) gene locus. J Bacteriol 181 3144 3154
18. IannelliFOggioniMRPozziG 2005 Sensor domain of histidine kinase ComD confers competence pherotype specificity in Streptoccoccus pneumoniae. FEMS Microbiol Lett 252 321 326
19. JohnsborgOKristiansenPEBlomqvistTHåvarsteinLS 2006 A Hydrophobic Patch in the Comptence-Stimulating Peptide, a Pneumococcal Competence Pheromone, Is Essential for Specificity and Biological Activity. J. Bacteriol 188 1744 1749
20. LauGWHaatajaSLonettoMKensitSEMarraA 2001 A functional genomic analysis of type 3 Streptococcus penumoniae virulence. Mol Microbiol 40 555 571
21. KowalkoJESebertME 2008 The Streptococcus pneumoniae competence regulatory system influences respiratory tract colonization. Infect Immun 76 3131 3140
22. IbrahimYMKerrARMcCluskeyJMitchellTJ 2004 Control of virulence by the two-component system CiaR/H is mediated via HtrA, a major virulence factor of Streptococcus pneumoniae. J Bacteriol 186 5258 5266
23. GuiralSMitchellTJMartinBClaverysJP 2005 Competence programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci USA 102 8710 8715
24. ClaverysJPMartinBHåvarsteinLS 2007 Competence-induced fratricide in streptococci. Mol Microbiol 64 1423 1433
25. SungCKLiHClaverysJPMorrisonDA 2001 An rpsL Cassette, Janus, for Gene Replacement through Negative Selection in Streptococcus pneumoniae. Appl Environ Microbiol 67 5190 5196
26. SungCKMorrisonDA 2005 Two Distinct Functions of ComW in Stabilization and Activation of the Alternative Sigma Factor ComX in Streptococcus pneumoniae. J Bacteriol 187 3052 3061
27. MorrisonDABakerMF 1978 Competence for genetic transformation in pneumococcus depends on synthesis of a small set of proteins. Nature 282 215 217
28. GriffithF 1922 The significance of pneumococcal types. J Hygiene 27 113 159
29. OttolenghiEMacLeodCM 1963 Genetic transformation among living pneumococci in the mouse. Proc Natl Acad Sci U S A 50 417 419
30. ConantJESawyerWD 1967 Transformation during mixed pneumococcal infection of mice. J Bact 93 1869 1875
31. Auerbach-RubinFOttolenghi-NightingaleE 1971 Effect of eathanol on the clearance of airborne pneumococci and the rate of pneumococcal transformation in the lung. Infect Immun 3 688 693
32. OggioniMRIannelliFRicciSChiavoliniDParigiR 2004 Antibacterial activity of a competence-stimulating peptide in experimental sepsis caused by Streptococcus pneumoniae. Antimicrob Agents Chemother 48 4725 4732
33. CoomaraswamyG 1996 Induction of genetic transformation in Streptococcus pneumoniae by a pheromone peptide and its synthetic analogues. Ph.D. thesis Chicago University of Illinois
34. HavarsteinLGMorrisonDA 1999 Quorum sensing and peptide pheromones in streptococcal competence for genetic transformation. DunnyGMWinansSC Cell-Cell Signaling in Bacteria Washington, D.C. American Society for Microbiology 9 26
35. OggioniMRTrappettiCKadiogluACassoneMIannelliF 2006 Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol Microbiol 61 1196 1210
36. LynchJP3rdZhanelGG 2009 Streptococcus pneumoniae: does antimicrobial resistance matter? Semin Respir Crit Care Med 30 210 238
37. JonesRNJacobsMRSaderHS 2010 Streptococcus pneumoniae resistance: implications for therapy of community-acquired bacterial pneumonia. Int J Antimicrob Agents 36 197 204
38. KlugmanKPLonksJR 2005 Hidden epidemic of macrolide-resistant pneumococci. Emerg Infect Dis 11 802 827
39. JacobsMR 2008 Antimicrobial-resistant Streptococcus pneumoniae: trends and management. Expert Rev Anti Infect Ther 6 619 635
40. PozziGIannelliFOggioniMRSantagatiMStefaniS 2004 Genetic elements carrying macrolide efflux genes in streptococci. Curr Drug Targets Infect Disord 4 203 206
41. WiddowsonCAKlugmanKP 1998 The molecular mechanisms of tetracycline resistance in the pneumococcus. Microb Drug Resist 4 79 84
42. CroucherNJWalkerDRomeroPLennardNPatersonGK 2009 Role of conjugative elements in the evolution of the multidrug-resistant pandemic clone Streptococcus pneumoniae Spain23F ST81. J Bacteriol 191 1480 1489
43. ShenXYangHYuSYaoKWangYYuanLYangY 2008 Macrolide-resistance mechanisms in Streptococcus pneumoniae isolates from Chinese children in association with genes of tetM and integrase of conjugative transposons 1545. Microb Drug Resist 14 155 161
44. DowsonCGHutchisonABranniganJAGeorgeRCHansmanD 1989 Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Nati Acad Sci USA 86 8842 8846
45. PrudhommeMAttaiechLSanchezGMartinBClaverysJP 2006 Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313 89 92
46. Ottolenghi-NightingaleE 1969 Spontaneously occurring bacterial transformations in mice. J Bacteriol 100 445 452
47. Ottolenghi-NightingaleE 1972 Competence of pneumococcal isolates and bacterial transformations in man. Infect Immun 6 785 92
48. DunnyGMLeonardBA 1997 Cell-cell communication in gram-positive bacteria. Annu Rev Microbiol 51 527 564
49. KleerebezemMQuadriLEKuipersOPde VosWM 1997 Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol 24 895 904
50. SturmeMHKleerebezemMNakayamaJAkkermansADVaughaEE 2002 Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Van Leeuwenhoek 81 233 243
51. LyonGJNovickRP 2004 Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 25 1389 1403
52. ThoendelMHorswillAR 2010 Biosynthesis of peptide signals in gram-positive bacteria. Adv Appl Microbiol 71 91 112
53. HåvarsteinLSHakenbeckRGaustadP 1997 Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J Bacteriol 179 6589 6594
54. KilianMPoulsenKBlomqvistTHåvarsteinLSBek-ThomsenM 2008 Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One 3 e2683
55. AveryOTMacLeodCMMcCartyM 1944 Studies on the chemical nature of the substance inducing transformation of pneumococcal types: inductions of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 79 137 158
56. OttolenghiEHotchkissRD 1960 Appearance of genetic transforming activity in pneumococcal cultures. Science 132 1257 1258
57. TettelinHNelsonKEPaulsenITEisenJAReadTD 2001 Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293 498 506
58. KausmallyLJohnsborgOLundeMKnutsenEHåvarsteinLS 2005 Choline-binding protein D (CbpD) in Streptococcus pneumoniae is essential for competence-induced cell lysis. J Bacteriol 187 4338 4345
59. ClaverysJ-PDintilhacAPestovaEVMartinBMorrisonDA 1995 Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis iin Streptococcus pneumoniae, using an ami test platform. Gene 164 123 128
60. EldholmVJohnsborgOStraumeDOhnstadHSBergKH 2010 CbpD is a murein hydrolase that requires a dual cell envelope binding specificity to kill target cells during fratricide. Mol Microbiol 76 905 917
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage EgressČlánek A Trigger Enzyme in : Impact of the Glycerophosphodiesterase GlpQ on Virulence and Gene ExpressionČlánek An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes byČlánek Th2-polarised PrP-specific Transgenic T-cells Confer Partial Protection against Murine ScrapieČlánek Alterations in the Transcriptome during Infection with West Nile, Dengue and Yellow Fever Viruses
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 9- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Unconventional Repertoire Profile Is Imprinted during Acute Chikungunya Infection for Natural Killer Cells Polarization toward Cytotoxicity
- Envelope Deglycosylation Enhances Antigenicity of HIV-1 gp41 Epitopes for Both Broad Neutralizing Antibodies and Their Unmutated Ancestor Antibodies
- Co-opts GBF1 and CERT to Acquire Host Sphingomyelin for Distinct Roles during Intracellular Development
- Nrf2, a PPARγ Alternative Pathway to Promote CD36 Expression on Inflammatory Macrophages: Implication for Malaria
- Robust Antigen Specific Th17 T Cell Response to Group A Streptococcus Is Dependent on IL-6 and Intranasal Route of Infection
- Targeting of a Chlamydial Protease Impedes Intracellular Bacterial Growth
- The Protease Cruzain Mediates Immune Evasion
- High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism
- Plague and Climate: Scales Matter
- Exhausted CD8 T Cells Downregulate the IL-18 Receptor and Become Unresponsive to Inflammatory Cytokines and Bacterial Co-infections
- Maturation-Induced Cloaking of Neutralization Epitopes on HIV-1 Particles
- Murine Gamma-herpesvirus Immortalization of Fetal Liver-Derived B Cells Requires both the Viral Cyclin D Homolog and Latency-Associated Nuclear Antigen
- Rapid and Efficient Clearance of Blood-borne Virus by Liver Sinusoidal Endothelium
- Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage Egress
- A Trigger Enzyme in : Impact of the Glycerophosphodiesterase GlpQ on Virulence and Gene Expression
- Strain Specific Resistance to Murine Scrapie Associated with a Naturally Occurring Human Prion Protein Polymorphism at Residue 171
- Development of a Transformation System for : Restoration of Glycogen Biosynthesis by Acquisition of a Plasmid Shuttle Vector
- Monalysin, a Novel -Pore-Forming Toxin from the Pathogen Contributes to Host Intestinal Damage and Lethality
- Host Phylogeny Determines Viral Persistence and Replication in Novel Hosts
- BC2L-C Is a Super Lectin with Dual Specificity and Proinflammatory Activity
- Expression of the RAE-1 Family of Stimulatory NK-Cell Ligands Requires Activation of the PI3K Pathway during Viral Infection and Transformation
- Structure of the Vesicular Stomatitis Virus N-P Complex
- HSV Infection Induces Production of ROS, which Potentiate Signaling from Pattern Recognition Receptors: Role for S-glutathionylation of TRAF3 and 6
- The Human Papillomavirus E6 Oncogene Represses a Cell Adhesion Pathway and Disrupts Focal Adhesion through Degradation of TAp63β upon Transformation
- Analysis of Behavior and Trafficking of Dendritic Cells within the Brain during Toxoplasmic Encephalitis
- Exposure to the Viral By-Product dsRNA or Coxsackievirus B5 Triggers Pancreatic Beta Cell Apoptosis via a Bim / Mcl-1 Imbalance
- Multidrug Resistant 2009 A/H1N1 Influenza Clinical Isolate with a Neuraminidase I223R Mutation Retains Its Virulence and Transmissibility in Ferrets
- Structure of Herpes Simplex Virus Glycoprotein D Bound to the Human Receptor Nectin-1
- Step-Wise Loss of Bacterial Flagellar Torsion Confers Progressive Phagocytic Evasion
- Complex Recombination Patterns Arising during Geminivirus Coinfections Preserve and Demarcate Biologically Important Intra-Genome Interaction Networks
- An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes by
- Non-Lytic, Actin-Based Exit of Intracellular Parasites from Intestinal Cells
- The Fecal Viral Flora of Wild Rodents
- The General Transcriptional Repressor Tup1 Is Required for Dimorphism and Virulence in a Fungal Plant Pathogen
- Interferon Regulatory Factor-1 (IRF-1) Shapes Both Innate and CD8 T Cell Immune Responses against West Nile Virus Infection
- A Small Non-Coding RNA Facilitates Bacterial Invasion and Intracellular Replication by Modulating the Expression of Virulence Factors
- Evaluating the Sensitivity of to Biotin Deprivation Using Regulated Gene Expression
- The Motility of a Human Parasite, , Is Regulated by a Novel Lysine Methyltransferase
- Phosphodiesterase-4 Inhibition Alters Gene Expression and Improves Isoniazid – Mediated Clearance of in Rabbit Lungs
- Restoration of IFNγR Subunit Assembly, IFNγ Signaling and Parasite Clearance in Infected Macrophages: Role of Membrane Cholesterol
- Protease ROM1 Is Important for Proper Formation of the Parasitophorous Vacuole
- The Regulated Secretory Pathway in CD4 T cells Contributes to Human Immunodeficiency Virus Type-1 Cell-to-Cell Spread at the Virological Synapse
- Rerouting of Host Lipids by Bacteria: Are You CERTain You Need a Vesicle?
- Transmission Characteristics of the 2009 H1N1 Influenza Pandemic: Comparison of 8 Southern Hemisphere Countries
- Th2-polarised PrP-specific Transgenic T-cells Confer Partial Protection against Murine Scrapie
- Sequential Bottlenecks Drive Viral Evolution in Early Acute Hepatitis C Virus Infection
- Genomic Insights into the Origin of Parasitism in the Emerging Plant Pathogen
- Genomic and Proteomic Analyses of the Fungus Provide Insights into Nematode-Trap Formation
- Influenza Virus Ribonucleoprotein Complexes Gain Preferential Access to Cellular Export Machinery through Chromatin Targeting
- Alterations in the Transcriptome during Infection with West Nile, Dengue and Yellow Fever Viruses
- Protease-Sensitive Conformers in Broad Spectrum of Distinct PrP Structures in Sporadic Creutzfeldt-Jakob Disease Are Indicator of Progression Rate
- Vaccinia Virus Protein C6 Is a Virulence Factor that Binds TBK-1 Adaptor Proteins and Inhibits Activation of IRF3 and IRF7
- c-di-AMP Is a New Second Messenger in with a Role in Controlling Cell Size and Envelope Stress
- Structural and Functional Studies on the Interaction of GspC and GspD in the Type II Secretion System
- APOBEC3A Is a Specific Inhibitor of the Early Phases of HIV-1 Infection in Myeloid Cells
- Impairment of Immunoproteasome Function by β5i/LMP7 Subunit Deficiency Results in Severe Enterovirus Myocarditis
- HTLV-1 Propels Thymic Human T Cell Development in “Human Immune System” Rag2 gamma c Mice
- Tri6 Is a Global Transcription Regulator in the Phytopathogen
- Exploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
- The Next Opportunity in Anti-Malaria Drug Discovery: The Liver Stage
- Significant Effects of Antiretroviral Therapy on Global Gene Expression in Brain Tissues of Patients with HIV-1-Associated Neurocognitive Disorders
- Inhibition of Competence Development, Horizontal Gene Transfer and Virulence in by a Modified Competence Stimulating Peptide
- A Novel Metal Transporter Mediating Manganese Export (MntX) Regulates the Mn to Fe Intracellular Ratio and Virulence
- Rhoptry Kinase ROP16 Activates STAT3 and STAT6 Resulting in Cytokine Inhibition and Arginase-1-Dependent Growth Control
- Hsp90 Governs Dispersion and Drug Resistance of Fungal Biofilms
- Secretion of Genome-Free Hepatitis B Virus – Single Strand Blocking Model for Virion Morphogenesis of Para-retrovirus
- A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence
- Membrane Remodeling by the Double-Barrel Scaffolding Protein of Poxvirus
- A Diverse Population of Molecular Type VGIII in Southern Californian HIV/AIDS Patients
- Disruption of TLR3 Signaling Due to Cleavage of TRIF by the Hepatitis A Virus Protease-Polymerase Processing Intermediate, 3CD
- Quantitative Analyses Reveal Calcium-dependent Phosphorylation Sites and Identifies a Novel Component of the Invasion Motor Complex
- Discovery of the First Insect Nidovirus, a Missing Evolutionary Link in the Emergence of the Largest RNA Virus Genomes
- Old World Arenaviruses Enter the Host Cell via the Multivesicular Body and Depend on the Endosomal Sorting Complex Required for Transport
- Exploits a Unique Repertoire of Type IV Secretion System Components for Pilus Assembly at the Bacteria-Host Cell Interface
- Recurrent Signature Patterns in HIV-1 B Clade Envelope Glycoproteins Associated with either Early or Chronic Infections
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- HTLV-1 Propels Thymic Human T Cell Development in “Human Immune System” Rag2 gamma c Mice
- Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage Egress
- Exploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
- A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



