-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaPto Kinase Binds Two Domains of AvrPtoB and Its Proximity to the Effector E3 Ligase Determines if It Evades Degradation and Activates Plant Immunity
Plant pathogenic bacteria inject effector proteins into plant cells to suppress immune responses and cause disease. The causal agent of bacterial speck, Pseudomonas syringae pv. tomato, is an important pathogen of tomato and a model system to study molecular plant-pathogen interactions. Here we report new insights into how the AvrPtoB effector can be recognized by the tomato kinase Pto to activate immunity. AvrPtoB is an active E3 ligase that is able to ubiquitinate host proteins and target them for degradation. The ability of Pto to resist ubiquitination and activate immunity has been attributed to its capacity to phosphorylate and inactivate the E3 ligase domain of AvrPtoB. Here we report that Pto can bind two distinct domains of AvrPtoB. Pto bound to the domain near the E3 ligase is degraded, whereas the distally bound Pto escapes ubiquitination. Furthermore, a kinase-inactive variant of Pto is fully capable of activating immunity in response to AvrPtoB, showing that proximity to the E3 ligase domain and not effector phosphorylation determines Pto recalcitrance to degradation. Our study provides further insight into the mechanism evolved by tomato to counteract a pathogenicity determinant of a bacterial pathogen, allowing it to activate an effective immune response.
Published in the journal: . PLoS Pathog 10(7): e32767. doi:10.1371/journal.ppat.1004227
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004227Summary
Plant pathogenic bacteria inject effector proteins into plant cells to suppress immune responses and cause disease. The causal agent of bacterial speck, Pseudomonas syringae pv. tomato, is an important pathogen of tomato and a model system to study molecular plant-pathogen interactions. Here we report new insights into how the AvrPtoB effector can be recognized by the tomato kinase Pto to activate immunity. AvrPtoB is an active E3 ligase that is able to ubiquitinate host proteins and target them for degradation. The ability of Pto to resist ubiquitination and activate immunity has been attributed to its capacity to phosphorylate and inactivate the E3 ligase domain of AvrPtoB. Here we report that Pto can bind two distinct domains of AvrPtoB. Pto bound to the domain near the E3 ligase is degraded, whereas the distally bound Pto escapes ubiquitination. Furthermore, a kinase-inactive variant of Pto is fully capable of activating immunity in response to AvrPtoB, showing that proximity to the E3 ligase domain and not effector phosphorylation determines Pto recalcitrance to degradation. Our study provides further insight into the mechanism evolved by tomato to counteract a pathogenicity determinant of a bacterial pathogen, allowing it to activate an effective immune response.
Introduction
In the perpetual evolutionary arms race between hosts and pathogens, plants evolved two layers of inducible defense to protect themselves from infection [1]. The first layer is now commonly referred to as pattern-triggered immunity (PTI). At its core are cell surface host receptors that detect common, highly conserved molecular features of microbes, referred to as microbe-associated molecular patterns. These receptors activate a relatively mild but effective defense response that includes the release of reactive oxygen species, changes in gene expression, and cell wall fortification [2], [3]. While this response is sufficient to prevent many potentially pathogenic microbes from successfully establishing an infection, adapted pathogens have evolved large arsenals of ‘effector’ proteins [4]–[6]. These effectors are typically delivered into the plant cell cytoplasm, where they interfere with immune signaling to subvert PTI [7]. The second plant immune response addresses this threat by monitoring for the presence of pathogen effectors and is consequently referred to as effector-triggered immunity (ETI) [8]–[10]. Because detection of an effector indicates the presence of a highly adapted and potentially devastating pathogen, the immune reaction is stronger and often includes localized cell death, referred to as the hypersensitive response [11]–[13]. Collectively, the ETI responses prevent the successful colonization by biotrophic or hemibiotrophic pathogens.
The interaction of Pseudomonas syringae pv. tomato DC3000 (Pst) with tomato is one of the best understood plant pathosystems [11], [14]. Pst delivers ∼28 effectors into the host cytoplasm via its type III secretion system in order to suppress PTI [4]. In response, certain wild relatives of tomato have evolved the capacity to recognize two of these effectors, AvrPto and AvrPtoB [15]. These proteins are unrelated, but both can be bound by the host Pto kinase which then acts with an NB-LRR protein, Prf, to trigger ETI [16]–[21]. Pto is a member of a small gene family in tomato and another family member, Fen, encodes a protein that is also able to bind AvrPtoB [22], [23]. However, the C-terminus of AvrPtoB contains a U-box type E3 ubiquitin ligase domain that marks Fen for degradation by the proteasome, thereby undermining Fen-mediated immunity [22], [24]–[27]. If the E3 ligase domain is incapacitated, Fen is able to activate ETI in conjunction with Prf [22].
Although Pto and Fen have 87% amino acid similarity, Fen is efficiently ubiquitinated by the AvrPtoB E3 ligase and degraded, whereas Pto is recalcitrant to ubiquitination and capable of activating ETI in response to binding AvrPtoB [22]. Understanding the molecular basis for this difference could shed light on an evolutionary step in the tomato-Pst interaction and also reveal an interesting mechanism underlying ETI. It has been reported recently that Pto is protected from AvrPtoB-mediated degradation because it has a significantly higher kinase activity than Fen [28]. This increased kinase activity was proposed to enable Pto to more efficiently phosphorylate AvrPtoB specifically at threonine-450 and this modification was reported to inactivate the AvrPtoB E3 ligase domain, allowing Pto to escape degradation [28].
A second important difference between Pto and Fen relates to the subdomains of AvrPtoB that they bind. A set of truncated versions of the effector was tested for interaction with both protein kinases, and only Pto was found to interact with an N-terminal fragment contained within amino acids 1–307 [22], [27], [29]. This region of AvrPtoB is also known to bind another tomato kinase, Bti9, thereby interfering with its role in PTI [30]. The structure of Pto in complex with this N-terminal binding site on AvrPtoB has been resolved by x-ray crystallography [17]. In contrast, Fen cannot interact with AvrPtoB1–307, but requires a longer fragment spanning amino acids 1-387 for interaction [22]. While Fen has so far eluded structural analysis, the additional C-terminal region of the effector necessary for interaction with Fen contains the binding site for the PTI co-receptor kinase BAK1, an important virulence target of AvrPtoB [25], [31].
A current model postulates that the AvrPtoB BAK1-interacting domain (BID) originally evolved as a virulence determinant to bind and suppress host kinases involved in PTI [15], [22]. Fen evolved as a decoy of these kinases, allowing it to interact with the BID (for clarity, here we will refer to this domain as the Fen-interacting domain, FID) and activate ETI [15]. The FID region of avrPtoB underwent an intragenic duplication creating a structurally similar N-terminal domain that diverged to target additional host protein kinases involved in PTI [25], [30]. The AvrPtoB E3 ligase domain was then acquired which effectively defeated Fen leading to disease susceptibility. However, multiple duplications of the Fen gene occurred in tomato and the protein encoded by one of the resulting genes, Pto, gained the ability to bind the N-terminal domain of AvrPtoB (i.e., the Pto-interacting domain, PID) and resist ubiquitination by the AvrPtoB E3 ligase. This stand-off appears to be the current stage in this plant-pathogen co-evolutionary arms race.
Here we investigate how Pto binds AvrPtoB and the mechanism by which this host kinase is able to resist ubiquitination by the AvrPtoB E3 ligase and activate the plant immune response.
Results
Pto binds two distinct domains of AvrPtoB, whereas Fen binds only an E3 ligase-proximal domain
The closely related kinases, Pto and Fen, from Solanum pimpinellifolium differ in their interactions with AvrPtoB [22], [29]. Pto interacts with the Pto-interacting domain (PID) contained within amino acids 1–307 of the effector (AvrPtoB1–307), whereas Fen interacts exclusively with the Fen-interacting domain (FID) spanning amino acids 307–387, present in the fragment AvrPtoB1–387 (Fig. 1A). To further examine these interactions, we subjected a number of AvrPtoB variants, with point mutations in the PID and FID domains, to yeast two-hybrid analyses with Fen and Pto (Fig. 1B). As reported previously, Pto interacted with AvrPtoB1–307, but was unable to interact with AvrPtoB1–307(F173A) [32]. Pto also interacted with AvrPtoB1–387, because this fragment contains the 1–307 region, but Fen interacted only with AvrPtoB1–387. Surprisingly, we found that Pto also interacted strongly with the AvrPtoB1–387(F173A) protein in which the PID has been inactivated. This indicated that Pto binds two distinct domains of AvrPtoB, the PID and FID, whereas Fen interacts only with the FID. To corroborate this finding, we included an AvrPtoB variant, AvrPtoB1–387(G325A) that is unable to interact with Fen. We found that the G325A substitution abolished the interaction of both Fen (no interaction of Fen with AvrPtoB1–387(G325A)) and Pto (no interaction of Pto with AvrPtoB1–387(F173A/G325A)) with the FID, but had no impact on the binding of Pto with the PID (strong interaction of Pto with AvrPtoB1–387(G325A)). These findings indicate that Fen interaction with AvrPtoB is limited to the FID, whereas Pto has the ability to bind both the FID and the PID.
Fig. 1. Pto, but not Fen, binds the PID of AvrPtoB and is recalcitrant to E3 ligase-mediated degradation. 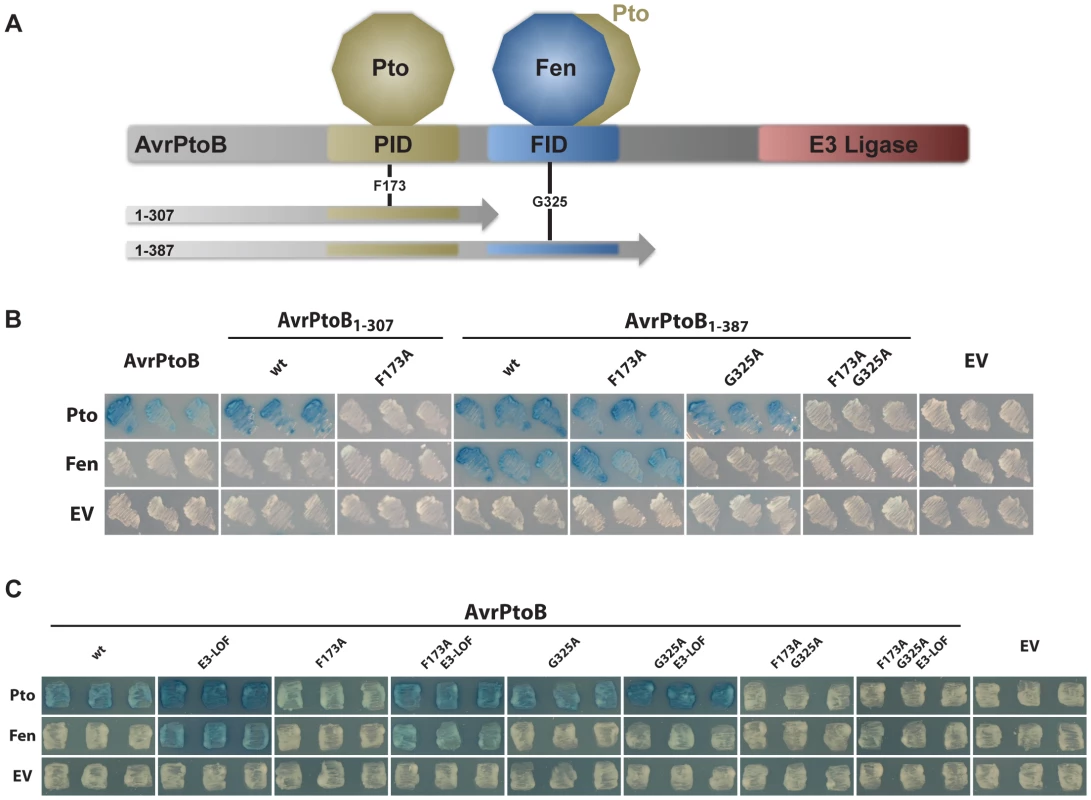
(A) Schematic of AvrPtoB functional domains and truncations used in this and previous publications [24]. PID, Pto-interacting domain [29]; FID, Fen-interacting domain (also known as the Bak1-interacting domain [25]). Position of amino acid substitutions, F173A and G325A, are shown. (B) Yeast two-hybrid analyses testing the interaction of tomato Pto and Fen kinases (in the bait vector) with different functional domains in AvrPtoB using truncated forms of the effector (in the prey vector). Blue patches indicate a positive interaction. Pto interacted with both the PID and the FID, whereas Fen exclusively bound the FID. wt, wild-type AvrPtoB in the truncation indicated; EV, empty vector. (C) Yeast two-hybrid analyses of the binding properties of tomato Pto and Fen kinases (in the bait vector) towards full-length AvrPtoB protein (in the prey vector). Blue patches indicate a positive interaction. Pto bound to the FID, proximal to the E3 ubiquitin ligase domain, was degraded in an E3-ligase dependent manner, similar to Fen. E3-LOF, AvrPtoB(F479A/F525A/P533A) has substitutions that abolish binding of the E2 conjugating enzyme and lacks E3 ligase activity [26]; EV, empty vector. Pto that binds to the E3 ligase-proximal FID is susceptible to degradation similar to Fen
To gain further insight into the interactions of Pto and Fen with AvrPtoB, we generated all combinations of amino acid substitutions in the FID and PID in full-length constructs that had an inactive E3 ligase (E3-LOF, having substitutions in the three E2-conjugating enzyme binding sites: F479A/F525A/P533A) (Fig. 1C). In the context of this full-length effector with an inactive E3 ligase, both Pto and Fen showed the same binding capabilities towards AvrPtoB as towards the truncated forms. Specifically, Pto could bind to both the PID and the FID, but Fen bound only to the FID. The presence of E3 ligase activity in the AvrPtoB C-terminus masked the interaction with Fen by marking the kinase for degradation, as described previously [22], [26]. Interestingly, if Pto was forced to bind exclusively to the FID by inclusion of the F173A substitution in the PID, this interaction was also masked by the E3 ligase activity, similar to Fen binding the same domain. These observations suggest that Pto is not intrinsically resistant to AvrPtoB-mediated degradation, but rather the proximity of its binding relative to the effector E3 ligase domain impacts whether or not Pto is ubiquitinated. We refer to this as the ‘proximity’ hypothesis.
Positioning of the PID directly adjacent to the E3 ligase domain renders Pto susceptible to AvrPtoB-mediated degradation
We tested the proximity hypothesis by generating a synthetic protein in which the PID (amino acids 121–200) was fused directly to the E3 ligase domain (amino acids 388–533) (Fig. 2). This PID fusion protein interacted strongly with Pto when the E3 ligase domain was inactivated. Significantly, this interaction was abrogated in the presence of an active E3 ligase domain, similar to what occurs when Fen or Pto bind the FID, indicating Pto is degraded when it binds a domain proximal to the E3 ligase domain. A Western blot showed that the PID fusion proteins were expressed as well as wild-type AvrPtoB (Fig. 2B) and the positive result of the interaction between Pto and PID:E3-LOF indicates that the kinase is expressed.
Fig. 2. AvrPtoB mediates Pto degradation if the kinase is positioned closer to the AvrPtoB E3 ligase. 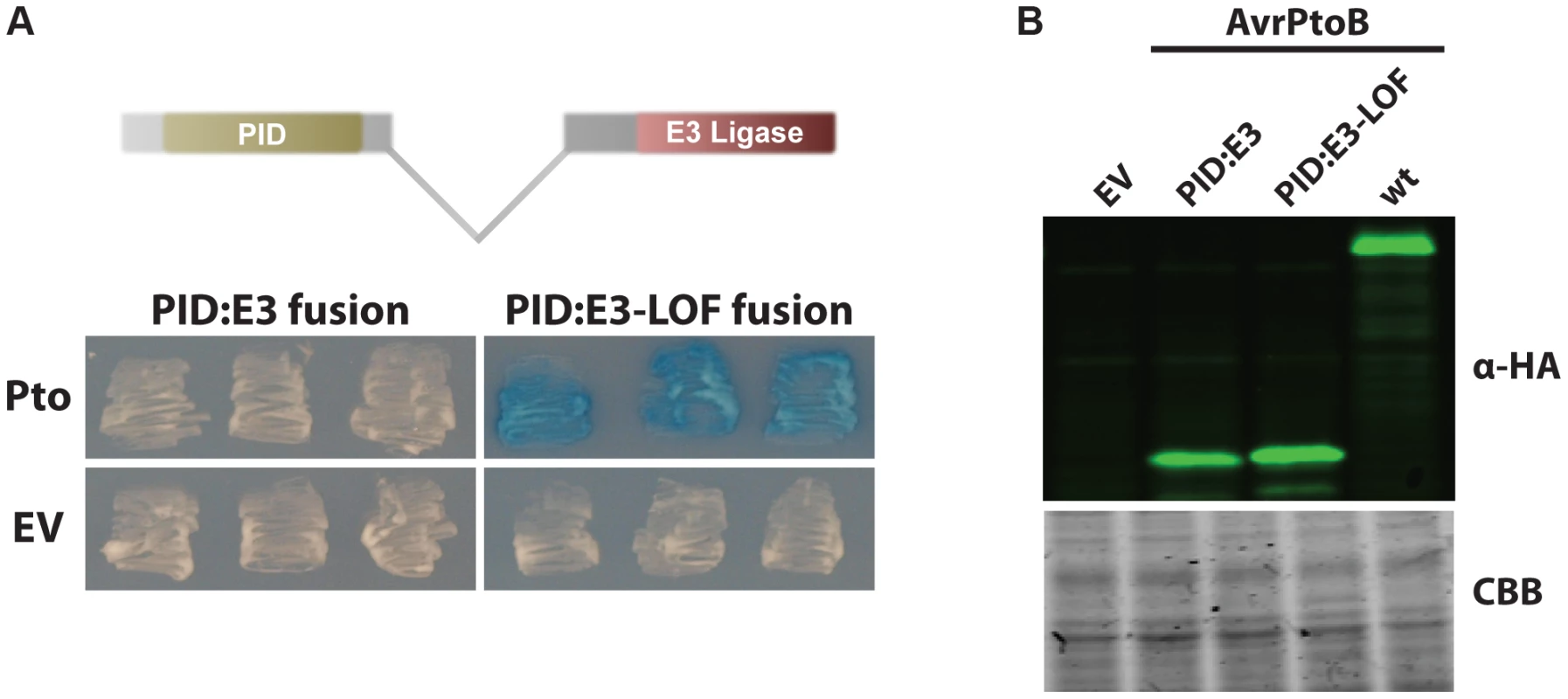
(A) The AvrPtoB PID was fused to the E3 ligase domain, cloned into the prey vector (see Methods for details) and tested for its interaction with Pto (in the bait vector). Blue patches indicate a positive interaction. Pto was degraded if it was in closer proximity to the E3 ligase domain as shown by the white patches. E3-LOF, AvrPtoB(F479A/F525A/P533A) lacks E3 ligase activity; EV, empty vector. (B) Expression levels of both fusion proteins and wild-type AvrPtoB are similar. Yeast cells grown in inductive medium were harvested, normalized by OD600, boiled in Laemmli buffer and proteins resolved by Western blotting. Fusion proteins were detected using ant-HA primary (clone 3F10, Roche, Indianapolis, IN, USA) and anti-rat-800 secondary (IRDye 800CW, LI-COR, Lincoln, NE, USA) antibodies and visualized using an Odyssey Scanner (LI-COR). CBB, Coomassie brilliant blue staining. Pto binding of the FID activates ETI in tomato and Nicotiana benthamiana leaves
We transformed Pst DC3000ΔavrPtoΔavrPtoB with the different variants of avrPtoB under control of a Pst hrp-inducible promoter and used these strains to infiltrate leaves of tomato plants that are either resistant (expressing Pto and Prf; Rio Grande-PtoR, RG-PtoR) or susceptible (carrying a mutation in Prf; Rio Grande-prf3, RG-prf3) (Fig. 3A). As observed previously, AvrPtoB(F173A) was unable to elicit Pto-mediated immunity [32]. However, in agreement with our yeast data, inactivation of the E3 ligase domain restored the immune response. A Fen knockout tomato line is not available thus making it impossible to test whether this reconstituted immune response is due to Pto or Fen (or both) binding to the FID. A tomato line lacking Prf (RG-prf3) and therefore deficient in both Fen - and Pto-mediated immunity was susceptible to all strains as expected (Fig 3A).
Fig. 3. Pto binds to the FID in plant cells and is degraded by activity of the AvrPtoB E3 ligase. 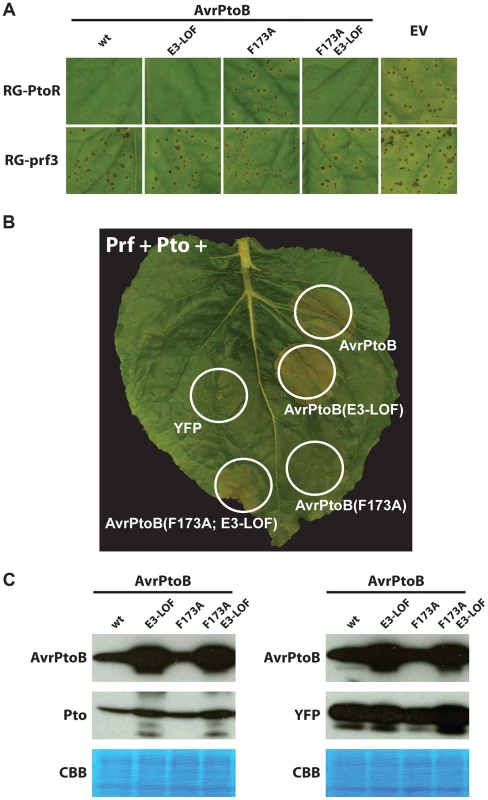
(A) The P. syringae pv. tomato strain DC3000ΔavrPtoΔavrPtoB was transformed with wild-type (wt) AvrPtoB or AvrPtoB variants under control of an Pst hrp promoter and infiltrated into leaves of Rio Grande (RG) tomatoes. Disease phenotypes were consistent with the observations in yeast two-hybrid analyses. RG-PtoR (Pto/Pto, Prf/Prf); RG-prf3 (Pto/Pto, prf3/prf3). RG-prf3 has a deletion in Prf that inactivates the Pto/Prf pathway. (B) Tomato Pto and Prf were co-expressed with different variants of AvrPtoB in a leaf of Nicotiana benthamiana by using Agrobacterium-mediated transient transformation. Pto activated ETI, as manifested by cell death with AvrPtoB(F173A)-E3-LOF, upon binding the FID but this response was repressed by the E3 ligase domain with no cell death visible with AvrPtoB(F173A), similar to Fen binding of the same domain. E3-LOF, AvrPtoB(F479A/F525A/P533A) lacks E3 ligase activity; YFP, yellow fluorescent protein control. (C) Abundance of Pto is decreased when it is co-expressed with AvrPtoB variants having an active E3 ligase domain. Using Agrobacterium-mediated transient transformation, tomato Pto was co-expressed with different variants of AvrPtoB in a leaf of a Prf-silenced Nicotiana benthamiana plant (to avoid the possible effect of cell death on protein abundance). Leaf samples were harvest 48 hrs after agroinfiltration and proteins were isolated for Western blot analysis. Similar experiments were done with yellow fluorescent protein (YFP) as a loading control. Proteins were detected using an anti-c-Myc-HRP antibody targeted to the epitope tag fused to each protein. Exposure was 1 min for AvrPtoB and Pto, and 10 seconds for YFP. CBB, Coomassie brilliant blue staining. To address the unavailability of tomato line lacking Fen, we employed Agrobacterium-mediated transient expression in Nicotiana benthamiana, a system that has been successfully used to investigate Pto - and Fen-mediated immunity [24], [27], [33]. It has been reported that one or more Pto family members in N. benthamiana recognize AvrPtoB1–387 and not AvrPtoB1-307 and can be degraded by the E3 ligase activity in AvrPtoB, analogous to degradation of Fen in tomato (referred to as the ‘Rsb’ phenotype; [22], [24]). However, when we tested new constructs expressing these truncations with C-terminal tags, we found unexpectedly that AvrPtoB1–307 was sufficient to activate Prf-dependent cell death in N. benthamiana (Fig. S1). Furthermore, the F173A mutation was sufficient to completely abolish recognition of E3 ligase-inactive AvrPtoB in N. benthamiana. We believe the earlier observations that N. benthamiana recognizes only AvrPtoB1–387 were due to low expression levels of the original AvrPtoB1–307 construct, which was not epitope-not tagged and for which expression levels were not tested [24].
The finding that the F173A substitution abolished the ability of AvrPtoB to elicit ETI-associated cell death in N. benthamiana gave us the opportunity to test whether Pto binding to the FID can activate ETI and, if so, whether this response can be suppressed by the AvrPtoB E3 ligase domain (Fig. 3B). It has previously been shown that tomato Pto can recognize full-length AvrPtoB in N. benthamiana if it is co-expressed with the tomato Prf gene [33]. In agreement with our yeast results, co-expression of Pto and Prf with avrPtoB(F173A/E3-LOF) caused ETI-associated cell death in N. benthamiana and this response was suppressed by the presence of a functional E3 ligase in AvrPtoB(F173A). A Western blot indicated that, as expected, Pto abundance was decreased in the presence of wild type AvrPtoB and AvrPtoB(F173A), but unaffected in the presence of the E3-LOF variants of AvrPtoB (Fig. 3C). Note that the E3-LOF variants of AvrPtoB are expressed at a higher level that the E3 ligase active versions (probably because AvrPtoB autoubiquitinates and is degraded). Nevertheless, even with this increased expression, AvrPtoB-E3-LOF variants do not cause a decrease in the abundance of Pto. These results demonstrate not only that Pto can bind the FID and trigger ETI, but also that the E3 ligase domain can inhibit this Pto-mediated response just as it inhibits cell death activated by Fen binding to the FID.
Pto recalcitrance to degradation does not depend on phosphorylation of T450 in AvrPtoB
It has been reported that Pto has greater kinase activity than Fen, allowing it to more effectively phosphorylate threonine-450 (T450) in AvrPtoB [28]. This phosphorylation was reported to inactivate the E3 ligase of the effector, explaining how Pto, but not Fen, resists degradation and activates an immune response. Our results suggest that the site at which Fen and Pto bind AvrPtoB relative to its E3 ligase domain has an impact on the fate of the kinase and consequently on disease resistance. These observations could support a model in which the ability of Pto to bind the PID in addition to the FID allows it to more effectively phosphorylate T450 of AvrPtoB. To test this hypothesis, we generated an AvrPtoB protein with a T450A substitution and tested its interaction with Pto in yeast (Fig. 4A). We expected one of two outcomes: 1) the T450A mutation disrupts E3 ligase activity similar to the published T450D substitution [28], allowing AvrPtoB(T450A) to interact with both Fen and Pto; or 2) the T450A substitution has no impact on E3 ligase activity but prevents Pto from phosphorylating and inactivating AvrPtoB, leading to degradation of Pto and evasion of Pto-mediated immunity in vivo. However, the outcome of this experiment did not agree with either expectation. Instead, AvrPtoB(T450A) was indistinguishable from wild-type AvrPtoB in that it interacted with Pto, but masked its interaction with Fen (Fig. 4A). This was puzzling as AvrPtoB(T450A) reportedly has no E3 ligase activity in vitro [28].
Fig. 4. Pto-mediated phosphorylation of T450 in AvrPtoB does not impact Pto recalcitrance to E3 ligase-mediated degradation. 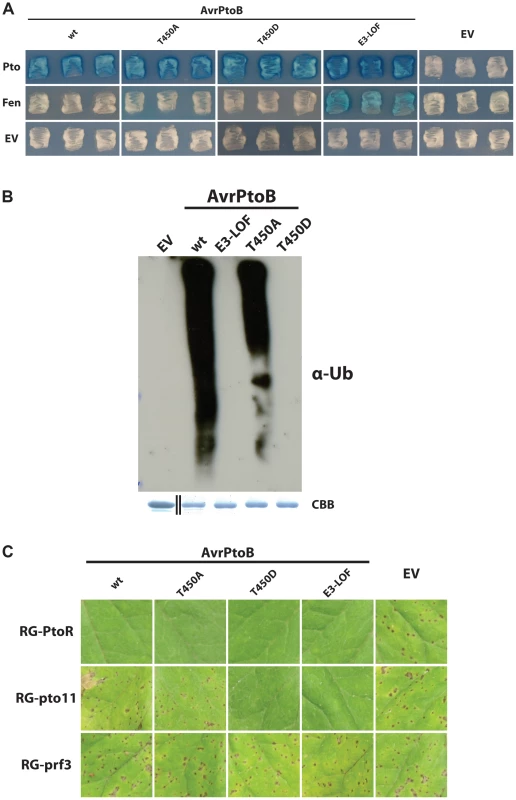
(A) Yeast two-hybrid analyses of the interaction of AvrPtoB(T450A) and AvrPtoB(T450D) with Pto and Fen. AvrPtoB or the variants shown (in the prey vector) were tested for their interaction with Pto and Fen. Blue patches indicate a positive interaction. Substitution of T450 in AvrPtoB by the non-phosphorylatable residue alanine had no impact on either the capability of the effector to degrade Fen or the recalcitrance of Pto to this degradation. AvrPtoB(T450D) interacted with Pto as expected based on results in (B) and (C) below. The reason for the lack of Fen interaction with AvrPtoB(T450D) is unknown as this interaction would be expected to occur based on (B) and evidently does occur in the plant-pathogen interaction as shown in (C). E3-LOF, AvrPtoB(F479A/F525A/P533A) lacks E3 ligase activity; EV, empty vector. (B) In vitro ubiquitination assay to determine ubiquitin ligase activities of different variants of AvrPtoB. GST-fusions of the effector were purified from E. coli and subjected to an in vitro ubiquitination assay. After Western blotting, poly-ubiquitin chains were detected using a monoclonal anti-ubiquitin antibody (α-Ub). In contrast to previous reports, AvrPtoB(T450A) retained significant E3 ubiquitin ligase activity. CBB, Coomassie Brilliant Blue. Black divider line in loading control panel demarcates a copy/paste border as GST has a much smaller mass than the fusion proteins. (C) The P. syringae pv. tomato strain DC3000ΔavrPtoΔavrPtoB was transformed with wild-type (wt) or variants of AvrPtoB under control of a Pst hrp promoter and infiltrated into Rio Grande (RG) tomatoes. AvrPtoB(T450D) was recognized both by Pto (no disease in RG-PtoR) and Fen (no disease in the absence of Pto in RG-pto11) and mimics the E3-LOF version of the effector. In contrast, AvrPtoB(T450A) behaved identical to wild-type AvrPtoB. Fen-mediated immunity is suppressed, but Pto can still resist degradation and confer immunity, demonstrating that phosphorylation of T450 in AvrPtoB is not required for Pto to resist degradation in vivo thus confirming the in vitro finding that AvrPtoB(T450A) has E3 ligase activity. RG-PtoR, Pto/Pto, Prf/Prf; RG-pto11, pto11/pto11, Prf/Prf; RG-prf3, Pto/Pto, prf3/prf3. RG-pto11 has a point mutation that produces a non-functional Pto protein. RG-prf3 has a deletion in Prf. To resolve this apparent contradiction, we repeated the experiment from Ntoukakis et al. [28] by purifying AvrPtoB(T450A) from E. coli and subjecting it to an in vitro E3 ligase assay (Fig. 4B). Consistent with our observation that AvrPtoB(T450A) can interact with Pto and cause degradation of Fen, this effector variant had only slightly reduced E3 ligase activity compared to wild-type AvrPtoB, whereas AvrPtoB(T450D) completely lacked E3 ligase activity.
This raised the question whether the slight reduction in E3 ligase activity caused by the T450A substitution is biologically significant or, as our results in yeast indicate, has no effect on the interplay between AvrPtoB and Pto. To make this distinction and stay as close to the in vivo situation as possible, we transformed Pst DC3000ΔavrPtoΔavrPtoB with different variants of avrPtoB under the control of a hrp-inducible promoter. The resulting strains were used to inoculate tomato leaves and disease or resistance outcomes were documented (Fig. 4C). RG-prf3 plants (lacking a functional Prf) served as a positive control for virulence; all of the strains were able to cause disease in this tomato line. RG-PtoR plants (expressing Pto and Prf) served to demonstrate the delivery of the AvrPtoB variants into the plant cell. In contrast to the observations with RG-prf3, none of the bacterial strains expressing any of the AvrPtoB variants, including AvrPtoB(T450A), caused disease in RG-PtoR plants. This result demonstrates that each of the AvrPtoB variants is successfully delivered into the plant cell cytoplasm, where Pto binding initiates ETI.
We next tested the bacterial strains on RG-pto11, a tomato line that has a nonfunctional Pto gene, rendering them susceptible to Pst expressing wild-type AvrPtoB (Fig. 4C). RG-pto11 plants do have a functional Fen, however, and are resistant to Pst with forms of AvrPtoB lacking E3 ligase activity [22], [24]. As expected, RG-pto11 tomatoes were susceptible to DC3000ΔavrPtoΔavrPtoB lacking AvrPtoB or expressing wild-type AvrPtoB, but resistant to strains with the E3 ligase-inactive variant AvrPtoB(E3-LOF). In agreement with both our yeast two-hybrid and E3 ligase assays as well as previously published data [28], RG-pto11 plants were also able to recognize AvrPtoB(T450D). However, RG-pto11 plants were unable to recognize AvrPtoB(T450A), indicating this effector variant is still able to suppress Fen-mediated ETI, which requires an active E3-ligase domain. This demonstrated not only that AvrPtoB(T450A) has E3 ligase activity, but also that the slight reduction in E3 ligase activity observed in the in vitro assays does not impact the plant-pathogen interaction. This result is consistent with our yeast two-hybrid analyses (Fig. 4A). Taken together with our observation that RG-PtoR plants are able to recognize AvrPtoB(T450A), this demonstrates that Pto phosphorylation of T450 in AvrPtoB is dispensable for Pto-mediated ETI triggered by AvrPtoB.
The relative in vitro kinase activities of Fen and Pto are dependent on buffer conditions
Ntoukakis et al. [28] reported a much reduced kinase activity for Fen relative to Pto and concluded this difference was a key factor in the ability of Pto, but not Fen, to phosphorylate T450 and thereby inactive the E3 ligase of AvrPtoB. These findings differ from earlier papers that described Pto and Fen as active kinases, with similar activities in vitro [34]–[37]. There are several differences in the protocols used in each case that might explain these discrepancies. In earlier assays from our laboratory, the kinases were expressed and purified as N-terminal maltose-binding protein (MBP)-fusions and the kinase assays were performed at pH 7.0 with 10 mM MnCl2 [34]–[37]. Ntoukakis et al. [28], however, purified C-terminally His-fusions and performed their assays at pH 7.5 with 10 mM MgCl2 and 1 mM MnCl2.
We reasoned that the different metal ions present in the assays were a likely explanation for the differences in kinase activities. Therefore we tested kinase activities with 10 mM MnCl2 and 10 mM MgCl2 at pH 7.5 and did observe higher kinase activity for Pto in the presence of 10 mM MnCl2 (Fig. S2A). However, we observed a brown discoloration of the kinase buffer at pH 7.5 in the presence of 10 mM MnCl2 (data not shown). We hypothesized this was due to a pH-dependent complex formation of Mn with Tris-HCl in solution. Indeed, such a pH-dependent complex formation has been described in the literature [38]. Further investigation showed that the observed discoloration increased with increasing pH and was dramatically enhanced upon addition of 1 mM DTT, as was used in all the kinase assays mentioned above, to the Tris buffers above pH 7.0 (Fig. S2B). This complex formation, together with the ten-fold lower MnCl2 concentration used by Ntoukakis et al. [28], could lead to a situation where the availability of Mn2+ is a limiting factor. To test whether the different kinase activities of Fen and Pto are due to limited availability of Mn at pH 7.5, we performed kinase assays with both proteins at pH 6.8. At pH 6.8 kinase activity of Fen was higher than that of Pto and this increased activity was dependent on the presence of Mn in the buffer (Fig. 5A).
Fig. 5. Fen has higher kinase activity than Pto and Pto(G50S) has little or no kinase activity. 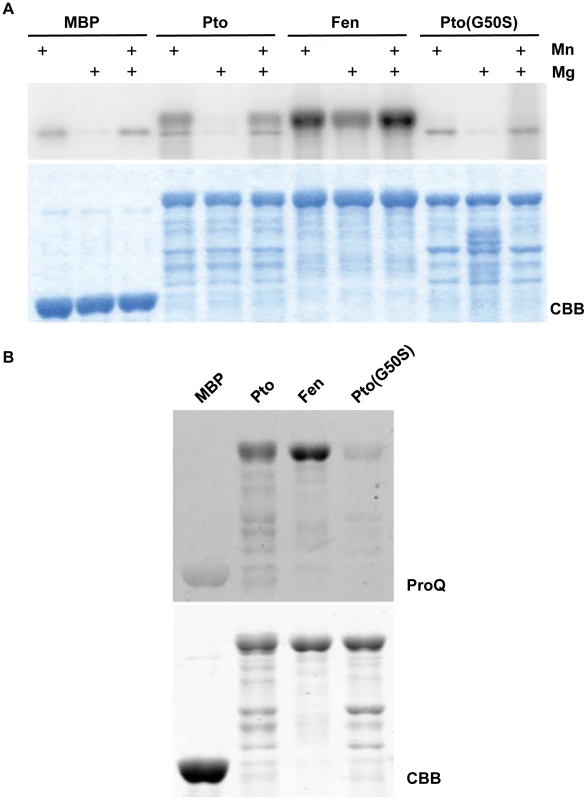
(A) In vitro kinase assay for tomato Pto, Fen, and Pto(G50S) at pH6.8. At this pH, no brown discoloration was visible upon addition of 10 mM MnCl2, indicating a better availability of Mn2+. Kinase buffers were supplemented with 10 mM MnCl2, 10 mM MgCl2 or 10 mM of each. Under these conditions, Fen showed a higher kinase activity than Pto and Pto(G50S) had little or no kinase activity. Coomassie Brilliant Blue (CBB) staining showed similar amounts of the kinases were present. (B) Phosphoprotein-specific ProQ staining of Pto, Fen and Pto(G50S) to assess their phosphorylation status in bacteria. Pto and Fen were expressed in E. coli, pulled down using MBP-agarose, resolved by SDS-PAGE and subjected to ProQ staining. Stronger staining of Fen indicates a higher autophosphorylation activity in situ. Pto(G50S) had little or no kinase activity in this in vivo assay. Coomassie Brilliant Blue (CBB) staining showed similar amounts of the kinases were present. The finding that the relative kinase activities of Fen and Pto are dependent on buffer pH and ion availability represents a general problem of in vitro experimentation. The chosen buffer conditions are to a certain degree arbitrary, and it is difficult to predict or replicate the exact in vivo conditions in the relevant subcellular locale. One way to address this limitation is to assess autophosphorylation activity of the kinases inside a living cell. Taylor et al. [39] recently published that many plant kinases, when expressed in E. coli, retain kinase activity and readily autophosphorylate. Furthermore, the level of this phosphorylation, as determined by using the phosphoamino acid stain Pro-Q Diamond, is indicative of the activity of the kinase under investigation [39]. We purified Fen and Pto from E. coli and assessed their phosphorylation status in vivo using Pro-Q staining (Fig. 5B). These experiments revealed that Fen autophosphorylation was approximately two - to four-fold higher than that of Pto. Together with our in vitro kinase assays in the presence of Mn, this indicates that under certain conditions Fen is a moderately more active kinase than Pto.
A Pto variant that lacks kinase activity still activates ETI in response to AvrPtoB
Our results suggest that Pto is not necessarily a more active kinase than Fen and we demonstrated that Pto phosphorylation of T450 is not required for its recalcitrance to AvrPtoB-mediated degradation. However, it is conceivable that in the absence of a phosphorylatable residue at position 450 and under the right conditions, Pto could phosphorylate neighboring amino acids which inactivate the E3 ligase domain. In fact, AvrPtoB has four serine residues within a distance of ten amino acids upstream or downstream of T450 that could serve as phospho-acceptors. We reasoned that the best way to exclude a role for Pto kinase activity in recalcitrance to AvrPtoB-mediated degradation would be to use a form of Pto that is kinase inactive, or at least less active than Fen under all conditions, but still capable of activating ETI. One Pto variant, Pto(G50S), was reported previously to retain the ability to activate ETI in response to AvrPto [33], [40]. G50 is an invariant residue in kinase subdomain I that plays a key role in binding ATP and its substitution would be expected to diminish or abolish kinase activity [41]. Indeed, when we purified Pto(G50S) from E. coli and subjected it to an in vitro assay we found that it had little or no kinase activity (Fig. 5A), a result confirmed by Pro-Q staining (Fig. 5B).
The transgenic tomato line expressing Pto(G50S) [40] is no longer available, so we utilized transient expression in N. benthamiana to test the ability of Pto(G50S) to activate ETI in response to AvrPtoB (Fig. 6). When co-expressed with tomato Prf, Pto(G50S) triggered ETI-associated cell death in response to AvrPtoB. In addition, as shown above for wild-type Pto (Fig. 3B), co-expression of Pto(G50S) with avrPtoB(F173A) did not activate ETI, but co-expression with an avrPtoB(F173A/E3-LOF) did. We conclude that with respect to its ability to recognize AvrPtoB and activate ETI, the kinase inactive Pto(G50S) is indistinguishable from wild-type Pto, demonstrating that Pto kinase activity is not required for Pto recalcitrance to AvrPtoB-mediated degradation.
Fig. 6. Pto and Fen kinase activities are dispensable for activation of AvrPtoB-elicited ETI. 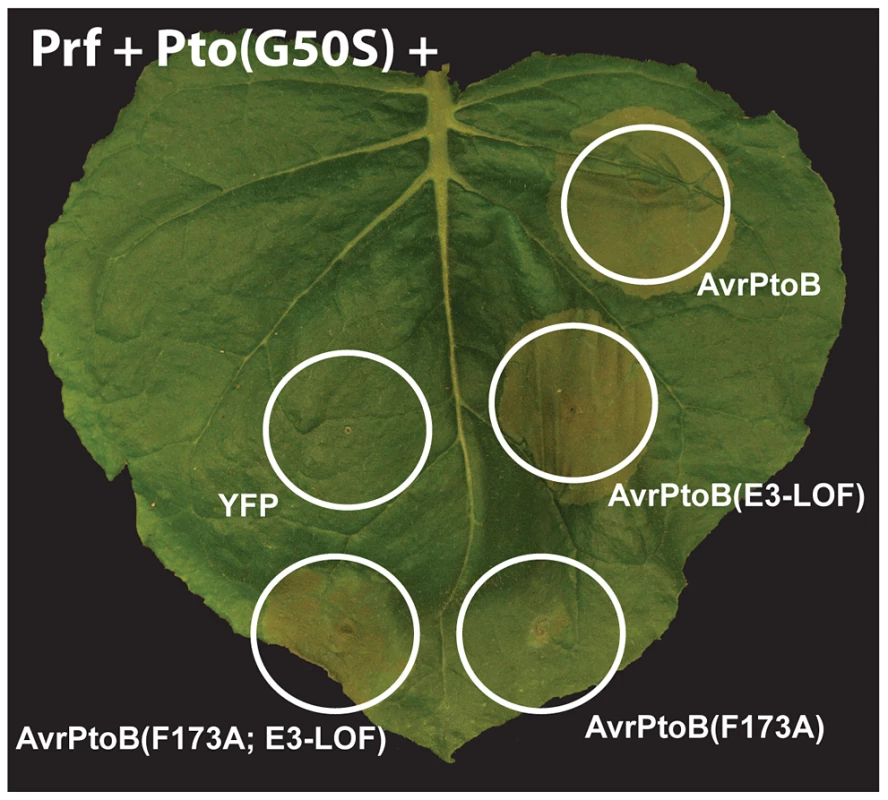
The Pto(G50S) variant which has little or no kinase activity (see Fig. 5BC) and tomato Prf were co-expressed with different variants of AvrPtoB in a leaf of Nicotiana benthamiana by Agrobacterium-mediated transient transformation. The response of Pto(G50S) to the AvrPtoB variants was indistinguishable from wild-type Pto in that it caused ETI-associated cell death upon binding the FID (cell deaths with AvrPtoB(F173A; E3-LOF)), but this response was repressed by the E3 ligase domain (no cell death by AvrPtoB(F173A)). Discussion
Testing variant forms of AvrPtoB for their interaction with Pto and Fen in a yeast two-hybrid system, revealed that Pto is capable of binding two distinct domains in the effector, both the N-terminal PID and the E3-ligase proximal FID, whereas Fen binds exclusively to the FID (Fig. S3). Furthermore, Pto that binds the FID is subjected to AvrPtoB-mediated degradation just as is Fen, showing that Pto is not intrinsically resistant to degradation. We also tested a construct in which the PID was fused directly to the E3 ligase domain and observed that Pto bound to this E3 ligase proximal PID was degraded. We initially interpreted these results as an indication that the position at which the kinases bind AvrPtoB relative to the E3 ligase domain might be the underlying cause for their apparent differential ability to phosphorylate T450 in AvrPtoB. We hypothesized that the unique ability of Pto to bind the more N-terminal PID allows Pto to efficiently phosphorylate AvrPtoB and thereby repress its E3 ligase activity. In contrast, Fen, which can only bind the more C-terminal FID, would not be in the optimal position and/or orientation to phosphorylate T450 and consequently be subject to ubiquitination and degradation.
When we tested the ability of Pto to activate ETI in response to AvrPtoB, we found that in the absence of E3 ligase activity Pto can trigger ETI upon binding either domain. However, only the Pto that binds the PID can activate ETI in the presence of an active AvrPtoB E3 ligase domain. Pto that binds the FID is targeted for degradation by the E3 ligase similar to Fen. These results are consistent with our yeast two-hybrid analyses and confirm that the binding sites of Pto and Fen on AvrPtoB determine the outcome. The presumed later evolution of the ability of Pto to bind the PID in addition to the FID allows it to escape AvrPtoB-mediated degradation and ultimately to trigger ETI.
To investigate the role of T450 phosphorylation in this process, we generated a version of AvrPtoB that can no longer be phosphorylated at this residue. Our results show that AvrPtoB(T450A) causes degradation of Fen in yeast and is capable of efficiently suppressing Fen-mediated ETI in tomato, properties that are identical to wild-type AvrPtoB. These observations were unexpected, as AvrPtoB(T450A) had previously been reported to lack E3 ligase activity in vitro [28]. When we repeated those experiments, we found that AvrPtoB(T450A) had only slightly reduced E3 ligase activity compared to wild-type AvrPtoB. The observation that AvrPtoB(T450A) still suppresses Fen-mediated ETI in tomato shows that this minor reduction is biologically insignificant. These results demonstrate that phosphorylation of T450, if that is what happens during the infection process, is not necessary for Pto-mediated ETI. The possibility remained that in the absence of a phosphorylatable residue at position 450, Pto might phosphorylate neighboring serine residues and thus interfere with E3 ligase activity.
The results described above demonstrate that the different binding characteristics of Pto and Fen underlie their differential vulnerability to degradation by AvrPtoB. They do not, however, exclude the possibility that a higher kinase activity of Pto suppresses AvrPtoB E3 ligase activity, assuming that other suitable phospho-acceptor residues exist in addition to T450. Unfortunately, there are inconsistent reports regarding the kinase activities of Pto and Fen. In some cases they were found to be similar [35]–[37], whereas Ntoukakis et al. [28] found Pto to be much more active than Fen, with Fen appearing almost kinase inactive. We found that the most likely explanation for these divergent results is the different buffer conditions under which the in vitro assays were performed, in particular the pH-dependent availability of Mn.
To assess the activity of both kinases independently of arbitrary buffer conditions, we employed the phospho-specific ProQ stain to determine autophosphorylation levels of both proteins when expressed in E. coli. Similar to the results obtained in vitro at pH 6.8, Fen appears to be a slightly more active kinase than Pto. At the very least, these results demonstrate that Pto does not have intrinsically higher kinase activity than Fen and that the relative activities are dependent on the buffer conditions used in the assay. We believe that the in vivo kinase activities observed in E. coli better reflect the kinase activities in plant cells, suggesting that Pto is not a more active kinase than Fen.
To conclusively address the importance of Pto kinase activity in its resistance to AvrPtoB-mediated degradation, we needed a version of Pto that is kinase inactive, but still capable of activating ETI in response to AvrPtoB. We took advantage of Pto(G50S) that confers resistance to AvrPto in tomato. Because G50 is an invariant residue in protein kinases and is involved in ATP binding, we suspected it would have little or no kinase activity. In fact, we were unable to detect any activity either in vitro or in E. coli. Pto(G50S) was able to activate programmed cell death in N. benthamiana in response to AvrPtoB as effectively as wild-type Pto. Together with Xiao et al. [40], this result demonstrates that Pto autophosphorylation is not required for activation of ETI in response to either AvrPto or AvrPtoB. The possibility remains that Pto transphosphorylation by another Pto family member as recently proposed [42] is required for the most efficient ETI response.
In retrospect, the model that Pto recalcitrance to AvrPtoB E3 ligase activity is due to phosphorylation of the E3 ligase by Pto is inconsistent with several observations. Specifically, expression of E3 ligase-deficient versions of AvrPtoB in N. benthamiana is known to trigger Fen-mediated ETI that can be suppressed by the active E3 ligase domain [22], [24]. In addition, co-expression of Pto with AvrPtoB in N. benthamiana does not alone activate ETI, but additionally requires additional expression of tomato Prf [33]. However, if Pto inactivates AvrPtoB E3 ligase activity by phosphorylating the E3 ligase domain, then co-expression of Pto with AvrPtoB in N. benthamiana should result in an E3 ligase-inactive AvrPtoB, which in turn should activate Fen-mediated ETI even in the absence of tomato Prf. However, this is not the case [33], which undermines the need to postulate that Pto phosphorylation of AvrPtoB plays a role in its recalcitrance to AvrPtoB E3 ligase activity [28].
In summary, our findings demonstrate that, in contrast to Fen, Pto evolved the capacity to not only bind the E3 ligase-proximal FID of AvrPtoB but additionally to bind the distal N-terminal PID. Interaction with this latter domain is what allows Pto to escape degradation and activate ETI in response to AvrPtoB. Because a kinase-inactive version of Pto is fully capable of activating ETI, phosphorylation of AvrPtoB to inactivate the E3 ligase activity is not a prerequisite for Pto-mediated immunity.
Materials and Methods
Plant material
Solanum lycopersicum cv. Rio-Grande (RG) and Nicotiana benthamiana (accession Nb-1; [43]) plants were grown in a greenhouse with 16 hr light/8 hr dark, 65% humidity and a temperature of 24°C during daylight and 22°C at night. Seeds were germinated on trays and plants were transferred to larger pots 2 weeks post germination.
Plant inoculations
Pseudomonas syringae pathovar tomato DC3000 with deletions in the avrPto and avrPtoB genes (DC3000ΔavrPtoΔavrPtoB; [44]) was transformed with different variants of AvrPtoB under control of a Pst hrp-inducible promoter in pCPP5372 [25]. Bacteria were grown on Kings B solid medium containing 100 µg/mL rifampicin and 10 µg/mL gentamicin at room temperature for 24 hr. The bacteria were scraped from the plates using sterile spatula, resuspended in sterile 10 mM MgCl2 and the OD600 was adjusted to 0.2 (corresponding to ∼5×108 CFU/mL). Serial dilutions in 10 mM MgCl2 were made to generate bacterial suspensions of 5×104 CFU/mL. Tomato plants were infiltrated by submersion of the aerial parts of the plant in the bacterial suspensions containing 0.002% Silwet L-77 and applying a vacuum of −80 kPa for 2 min. Plants were allowed to dry lying on their side and transferred into a growth chamber. Disease development was monitored for up to one week after infiltration.
Cloning
All enzymes and reagents used for cloning were purchased from New England Biolabs (Ipswich, MA, USA) unless otherwise noted. For Gateway entry vectors, the open-reading frames (ORFs) were amplified with or without a stop codon using Phusion DNA polymerase and blunt-end ligated into the SmaI site of pJM51 or pJLSmart. Following sequence confirmation, recombination reactions were performed using Gateway LR Clonase II (Life Technologies, Carlsbad, CA, USA) following the manufacturer's instructions to transfer the ORFs into Gateway compatible destination vectors. Recombination sites were confirmed by sequencing.
The yeast two-hybrid vectors used in this study (pEG202 and pJG4-5) are not Gateway compatible. Consequently, ORFs for expression in yeast were amplified using gene-specific primers adding EcoRI sites at the 5′ and 3′ ends and the PCR products were digested with EcoRI and ligated into pEG202 or pJG4-5. Orientation of the insert was confirmed by colony PCR, and the complete ORF of the inserted gene was confirmed by sequencing.
In vitro mutagenesis was performed following standard protocols (Stratagene QuikChange site directed mutagenesis kit). In brief, complementary oligos were designed for both strands containing the desired nucleotide changes flanked by at least 15 nucleotides perfectly matching the template DNA. These oligos were used in a 15 - to 18-cycle PCR with Phusion DNA polymerase and 50 ng of a vector containing the original (unchanged) DNA sequence. The PCR product was subjected to a DpnI digest to remove template plasmid and 2–5 µl of the resultant solution were used to transform E. coli DH5α.
Fusion protein ORFs were generated by fusion PCRs. First, PCR reactions were performed using oligos that added 15–25 nt of additional sequence homologous to the ‘other’ side of the desired fusion border to the 3′ (for the N-terminal fusion part) or 5′ (for the C-terminal fusion part) end of the PCR product. These PCR products were purified and used as templates in a PCR using only the oligos corresponding to the 5′ (forward oligo) and 3′ (reverse oligo) of the desired final fusion ORF, generating the full length fusion. Full-length PCR products were ligated into the different plasmids as described above and confirmed by sequencing.
Details of oligonucleotides (Table S1), vectors (Table S2), and constructs (Table S3) used in this work are provided in the supplemental information. Complete sequences as well as Gene Construction Kit (GCK) format vector maps of all constructs generated in this work are available upon request.
In vitro ubiquitin ligase assay
Ubiquitin ligase assays were performed as described previously [26]. In brief, 1 µg of purified GST:AvrPtoB was added to a solution containing 50 nM E1 (UBE1), 100 nM E2 (UbcH5a), 10 µg Ubiquitin, 2 mM DTT, 5 mM ATP, 5 mM MgCl2 and 50 mM Tris at pH 7.5 in a total volume of 30 µl for 2 hrs at 30°C. Reactions were stopped by the addition of 30 µl 2× reducing Laemmli buffer and boiling for 5 min. E1 (#E-304), E2 (#E2-616) and ubiquitin (#U-100) were purchased from Boston Biochemical (Cambridge, MA, USA). Proteins were resolved by SDS-PAGE, transferred to PVDF membranes by Western blotting and detected using monoclonal mouse anti-ubiquitin antibodies (clone P4D1, SC-8017, Santa Cruz Biotechnology, Santa Cruz, CA, USA) at a dilution of 1/5000.
In vitro kinase assays
Kinase assays were performed as described previously [35] and as elaborated upon in the text. Tris-HCl (50 mM) at either pH 6.8 or 7.5 and 10 mM of either MnCl2, MgCl2 or both were used in the buffers to test for the impact of buffer conditions on kinase activities.
Pro-Q phospho-protein staining
Proteins were purified from E. coli and resolved by SDS-PAGE. Pro-Q staining and Coomassie Silver counter-staining were performed exactly as described by Taylor et al. 2013 [39]. Pro-Q was obtained from Invitrogen Co. (Eugene, OR)
Yeast two-hybrid assays
The LexA yeast two-hybrid system was used to investigate the interactions of different forms of AvrPtoB with Pto and Fen [22], [24]. ORFs were inserted into the EcoRI sites of pEG202 (bait) or pJG4-5 (prey) vectors and introduced into Saccharomyces cerevisiae strain EGY48 containing the reporter plasmid pSH18-34. Transformations were plated onto CM (minus uracil, histidine, tryptophan, UHW) dropout medium containing glucose and single colonies of primary transformants were replica-plated onto fresh CM –UHW/glucose master plates and grown overnight. Subsequently, these colonies were replica-plated onto inductive CM –UHW plates containing raffinose/galactose and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) and staining was monitored for 24 to 48 hrs.
Agrobacterium-mediated transient protein expression
Agrobacterium tumefaciens strain GV3101 with helper plasmid pMP90 was transformed with constructs containing the genes to be expressed under control of a CaMV 35S promoter using electroporation. Single colonies were selected and the presence of the correct transgene confirmed by colony PCR. Agrobacteria were grown as a lawn on Luria Bertani medium plates containing appropriate antibiotics for 24 hr at 30°C. Bacteria were scraped from the plates using a sterile spatula and resuspended in sterile infiltration medium (10 mM MgCl2, 50 mM MES pH 5.6, 500 µM aceto-syringone) and the OD600 determined. Based on the OD, aliquots of the solutions corresponding to a desired density in a desired volume were transferred to fresh tubes, pelleted by centrifugation (2000 rcf, 10 min, RT), decanted and the pellets resuspended in the correct volume of fresh buffer.
Leaves of 4 to 6-week old N. benthamiana plants were infiltrated with bacterial suspensions containing an OD600 of 0.3 for each construct using a needle-less syringe. Plants were transferred to a growth chamber shelf with no lights and surrounded by shade cloth after infiltration. For analysis of protein expression by Western blotting, samples were collected 48 hr after infiltration. Cell death in the infiltrated areas was monitored for up to one week after infiltration; cell death typically became macroscopically visible 48 hr after infiltration and was completely developed 5 d after infiltration.
Supporting Information
Zdroje
1. BollerT, HeSY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324 : 742–744.
2. MonaghanJ, ZipfelC (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol 15 : 349–357.
3. RosliHG, ZhengY, PomboMA, ZhongS, BombarelyA, et al. (2013) Transcriptomics-based screen for genes induced by flagellin and repressed by pathogen effectors identifies a cell wall-associated kinase involved in plant immunity. Genome Biology 14: R139.
4. LindebergM, CunnacS, CollmerA (2012) Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol 20 : 199–208.
5. CunnacS, ChakravarthyS, KvitkoBH, RussellAB, MartinGB, et al. (2011) Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc Natl Acad Sci U S A 108 : 2975–2980.
6. BaltrusDA, NishimuraMT, RomanchukA, ChangJH, MukhtarMS, et al. (2011) Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog 7: e1002132.
7. FengF, ZhouJM (2012) Plant-bacterial pathogen interactions mediated by type III effectors. Curr Opin Plant Biol 15 : 469–476.
8. MoffettP (2009) Mechanisms of recognition in dominant R gene-mediated resistance. Advances Virus Res 75 : 1–33.
9. MaekawaT, KuferTA, Schulze-LefertP (2011) NLR functions in plant and animal immune systems: so far and yet so close. Nature Immunol 12 : 817–826.
10. DoddsPN, RathjenJP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11 : 539–548.
11. OhCS, MartinGB (2011) Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci 16 : 132–140.
12. CaplanJ, PadmanabhanM, Dinesh-KumarSP (2008) Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe 3 : 126–135.
13. TsudaK, SatoM, StoddardT, GlazebrookJ, KatagiriF (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772.
14. PedleyKF, MartinGB (2003) Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annual Review of Phytopathology 41 : 215–243.
15. Martin GB (2012) Suppression and activation of the plant immune system by Pseudomonas syringae effectors AvrPto and AvrPtoB. In: Martin F, Kamoun S, editors.Effectors in Plant-Microbe Interactions: Wiley-Blackwell. pp. 123–154.
16. XingW, ZouY, LiuQ, LiuJ, LuoX, et al. (2007) The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature 449 : 243–247.
17. DongJ, XiaoF, FanF, GuL, CangH, et al. (2009) Crystal structure of the complex between Pseudomonas effector AvrPtoB and the tomato Pto kinase reveals both a shared and a unique interface compared with AvrPto-Pto. Plant Cell 21 : 1846–1859.
18. GutierrezJR, BalmuthAL, NtoukakisV, MucynTS, Gimenez-IbanezS, et al. (2010) Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J 61 : 507–518.
19. MucynTS, WuAJ, BalmuthAL, ArastehJM, RathjenJP (2009) Regulation of tomato Prf by Pto-like protein kinases. Mol Plant Microbe Interact 22 : 391–401.
20. LinNC, MartinGB (2007) Pto - and Prf-mediated recognition of AvrPto and AvrPtoB restricts the ability of diverse Pseudomonas syringae pathovars to infect tomato. Mol Plant Microbe Interact 20 : 806–815.
21. SalmeronJM, OldroydGED, RommensCMT, ScofieldSR, KimH-S, et al. (1996) Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86 : 123–133.
22. RosebrockTR, ZengL, BradyJJ, AbramovitchRB, XiaoF, et al. (2007) A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448 : 370–374.
23. MartinGB, BrommonschenkelSH, ChunwongseJ, FraryA, GanalMW, et al. (1993) Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262 : 1432–1436.
24. AbramovitchRB, KimY-J, ChenS, DickmanMB, MartinGB (2003) Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J 22 : 60–69.
25. ChengW, MunkvoldKR, GaoH, MathieuJ, SchwizerS, et al. (2011) Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase-interacting domains in a type III effector. Cell Host Microbe 10 : 616–626.
26. JanjusevicR, AbramovitchRB, MartinGB, StebbinsCE (2006) A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311 : 222–226.
27. AbramovitchRB, JanjusevicR, StebbinsCE, MartinGB (2006) Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc Natl Acad Sci USA 103 : 2851–2856.
28. NtoukakisV, MucynTS, Gimenez-IbanezS, ChapmanHC, GutierrezJR, et al. (2009) Host inhibition of a bacterial virulence effector triggers immunity to infection. Science 324 : 784–787.
29. XiaoF, HeP, AbramovitchRB, DawsonJE, NicholsonLK, et al. (2007) The N-terminal region of Pseudomonas type III effector AvrPtoB elicits Pto-dependent immunity and has two distinct virulence determinants. Plant J 52 : 595–614.
30. ZengL, VelasquezAC, MunkvoldKR, ZhangJ, MartinGB (2012) A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J 69 : 92–103.
31. ShanL, HeP, LiJ, HeeseA, PeckSC, et al. (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4 : 17–27.
32. XiaoF, GiavaliscoP, MartinGB (2007) Pseudomonas syringae type III effector AvrPtoB is phosphorylated in plant cells on serine 258, promoting its virulence activity. J Biol Chem 282 : 30737–30744.
33. MucynTS, ClementeA, AndriotisVM, BalmuthAL, OldroydGE, et al. (2006) The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 18 : 2792–2806.
34. LohY-T, MartinGB (1995) The disease resistance gene Pto and the fenthion-sensitivity gene Fen encode closely related, functional protein kinases. Proc Natl Acad Sci USA 92 : 4181–4184.
35. LohY-T, MartinGB (1995) The Pto bacterial resistance gene and the Fen insecticide sensitivity gene encode functional protein kinases with serine/threonine specificity. Plant Physiol 108 : 1735–1739.
36. ZhouJ, LohYT, BressanRA, MartinGB (1995) The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 83 : 925–935.
37. JiaY, LohY-T, ZhouJ, MartinGB (1997) Alleles of Pto and Fen occur in bacterial speck-susceptible and fenthion-insensitive tomato and encode active protein kinases. Plant Cell 9 : 61–73.
38. FischerBE, HaringUK, TriboletR, SigelH (1979) Metal ion/buffer interactions. Stability of binary and ternary complexes containing 2-amino-2(hydroxymethyl)-1,3-propanediol (Tris) and adenosine 5′-triphosphate (ATP). Eur J Biochem 94 : 523–530.
39. TaylorI, SeitzK, BennewitzS, WalkerJC (2013) A simple in vitro method to measure autophosphorylation of protein kinases. Plant Methods 9 : 22.
40. XiaoF, LuM, LiJ, ZhaoT, YiSY, et al. (2003) Pto mutants differentially activate Prf-dependent, avrPto-independent resistance and gene-for-gene resistance. Plant Physiol 131 : 1239–1249.
41. Hanks SK, Hunter T (1995) The eukaryotic protein kinase superfamily. In: Hardie G, Hanks S, editors.The Protein Kinase Facts Book: Protein Serine Kinases.San Diego, CA: Harcourt Brace & Co. pp. 7–47.
42. NtoukakisV, BalmuthAL, MucynTS, GutierrezJR, JonesAM, et al. (2013) The tomato Prf complex is a molecular trap for bacterial effectors based on Pto transphosphorylation. PLoS Pathog 9: e1003123.
43. BombarelyA, RosliHG, VrebalovJ, MoffettP, MuellerLA, et al. (2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant-Microbe Interact 25 : 1523–1530.
44. LinNC, MartinGB (2005) An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol Plant Microbe Interact 18 : 43–51.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Molecular and Cellular Mechanisms of KSHV Oncogenesis of Kaposi's Sarcoma Associated with HIV/AIDSČlánek The Semen Microbiome and Its Relationship with Local Immunology and Viral Load in HIV InfectionČlánek Peptidoglycan Recognition Proteins Kill Bacteria by Inducing Oxidative, Thiol, and Metal Stress
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 7- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Bacteriophages as Vehicles for Antibiotic Resistance Genes in the Environment
- Helminth Infections, Type-2 Immune Response, and Metabolic Syndrome
- Defensins and Viral Infection: Dispelling Common Misconceptions
- Holobiont–Holobiont Interactions: Redefining Host–Parasite Interactions
- The Wide World of Ribosomally Encoded Bacterial Peptides
- Microbial Egress: A Hitchhiker's Guide to Freedom
- Molecular and Cellular Mechanisms of KSHV Oncogenesis of Kaposi's Sarcoma Associated with HIV/AIDS
- HIV-1 Capture and Transmission by Dendritic Cells: The Role of Viral Glycolipids and the Cellular Receptor Siglec-1
- Tetherin Can Restrict Cell-Free and Cell-Cell Transmission of HIV from Primary Macrophages to T Cells
- The Frustrated Host Response to Is Bypassed by MyD88-Dependent Translation of Pro-inflammatory Cytokines
- Larger Mammalian Body Size Leads to Lower Retroviral Activity
- The Semen Microbiome and Its Relationship with Local Immunology and Viral Load in HIV Infection
- Lytic Gene Expression Is Frequent in HSV-1 Latent Infection and Correlates with the Engagement of a Cell-Intrinsic Transcriptional Response
- Phase Variation of Poly-N-Acetylglucosamine Expression in
- A Screen of Mutants Reveals Important Roles for Dot/Icm Effectors and Host Autophagy in Vacuole Biogenesis
- Structure of the Trehalose-6-phosphate Phosphatase from Reveals Key Design Principles for Anthelmintic Drugs
- The Impact of Juvenile Coxsackievirus Infection on Cardiac Progenitor Cells and Postnatal Heart Development
- Vertical Transmission Selects for Reduced Virulence in a Plant Virus and for Increased Resistance in the Host
- Characterization of the Largest Effector Gene Cluster of
- Novel Drosophila Viruses Encode Host-Specific Suppressors of RNAi
- Pto Kinase Binds Two Domains of AvrPtoB and Its Proximity to the Effector E3 Ligase Determines if It Evades Degradation and Activates Plant Immunity
- Genetic Analysis of Tropism Using a Naturally Attenuated Cutaneous Strain
- Plasmacytoid Dendritic Cells Suppress HIV-1 Replication but Contribute to HIV-1 Induced Immunopathogenesis in Humanized Mice
- A Novel Mouse Model of Gastroenteritis Reveals Key Pro-inflammatory and Tissue Protective Roles for Toll-like Receptor Signaling during Infection
- Pathogenicity of Is Expressed by Regulating Metabolic Thresholds of the Host Macrophage
- BCKDH: The Missing Link in Apicomplexan Mitochondrial Metabolism Is Required for Full Virulence of and
- Independent Bottlenecks Characterize Colonization of Systemic Compartments and Gut Lymphoid Tissue by
- Peptidoglycan Recognition Proteins Kill Bacteria by Inducing Oxidative, Thiol, and Metal Stress
- G3BP1, G3BP2 and CAPRIN1 Are Required for Translation of Interferon Stimulated mRNAs and Are Targeted by a Dengue Virus Non-coding RNA
- Cytolethal Distending Toxins Require Components of the ER-Associated Degradation Pathway for Host Cell Entry
- The Machinery at Endoplasmic Reticulum-Plasma Membrane Contact Sites Contributes to Spatial Regulation of Multiple Effector Proteins
- Arabidopsis LIP5, a Positive Regulator of Multivesicular Body Biogenesis, Is a Critical Target of Pathogen-Responsive MAPK Cascade in Plant Basal Defense
- Plant Surface Cues Prime for Biotrophic Development
- Real-Time Imaging Reveals the Dynamics of Leukocyte Behaviour during Experimental Cerebral Malaria Pathogenesis
- The CD27L and CTP1L Endolysins Targeting Contain a Built-in Trigger and Release Factor
- cGMP and NHR Signaling Co-regulate Expression of Insulin-Like Peptides and Developmental Activation of Infective Larvae in
- Systemic Hematogenous Maintenance of Memory Inflation by MCMV Infection
- Strain-Specific Variation of the Decorin-Binding Adhesin DbpA Influences the Tissue Tropism of the Lyme Disease Spirochete
- Distinct Lipid A Moieties Contribute to Pathogen-Induced Site-Specific Vascular Inflammation
- Serovar Typhi Conceals the Invasion-Associated Type Three Secretion System from the Innate Immune System by Gene Regulation
- LANA Binds to Multiple Active Viral and Cellular Promoters and Associates with the H3K4Methyltransferase hSET1 Complex
- A Molecularly Cloned, Live-Attenuated Japanese Encephalitis Vaccine SA-14-2 Virus: A Conserved Single Amino Acid in the Hairpin of the Viral E Glycoprotein Determines Neurovirulence in Mice
- Illuminating Fungal Infections with Bioluminescence
- Comparative Genomics of Plant Fungal Pathogens: The - Paradigm
- Motility and Chemotaxis Mediate the Preferential Colonization of Gastric Injury Sites by
- Widespread Sequence Variations in VAMP1 across Vertebrates Suggest a Potential Selective Pressure from Botulinum Neurotoxins
- An Immunity-Triggering Effector from the Barley Smut Fungus Resides in an Ustilaginaceae-Specific Cluster Bearing Signs of Transposable Element-Assisted Evolution
- Establishment of Murine Gammaherpesvirus Latency in B Cells Is Not a Stochastic Event
- Oncogenic Herpesvirus KSHV Hijacks BMP-Smad1-Id Signaling to Promote Tumorigenesis
- Human APOBEC3 Induced Mutation of Human Immunodeficiency Virus Type-1 Contributes to Adaptation and Evolution in Natural Infection
- Innate Immune Responses and Rapid Control of Inflammation in African Green Monkeys Treated or Not with Interferon-Alpha during Primary SIVagm Infection
- Chitin-Degrading Protein CBP49 Is a Key Virulence Factor in American Foulbrood of Honey Bees
- Influenza A Virus Host Shutoff Disables Antiviral Stress-Induced Translation Arrest
- Nsp9 and Nsp10 Contribute to the Fatal Virulence of Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus Emerging in China
- Pulmonary Infection with Hypervirulent Mycobacteria Reveals a Crucial Role for the P2X7 Receptor in Aggressive Forms of Tuberculosis
- Syk Signaling in Dendritic Cells Orchestrates Innate Resistance to Systemic Fungal Infection
- A Repetitive DNA Element Regulates Expression of the Sialic Acid Binding Adhesin by a Rheostat-like Mechanism
- T-bet and Eomes Are Differentially Linked to the Exhausted Phenotype of CD8+ T Cells in HIV Infection
- Israeli Acute Paralysis Virus: Epidemiology, Pathogenesis and Implications for Honey Bee Health
- Influence of ND10 Components on Epigenetic Determinants of Early KSHV Latency Establishment
- Antibody to gp41 MPER Alters Functional Properties of HIV-1 Env without Complete Neutralization
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Molecular and Cellular Mechanisms of KSHV Oncogenesis of Kaposi's Sarcoma Associated with HIV/AIDS
- Holobiont–Holobiont Interactions: Redefining Host–Parasite Interactions
- BCKDH: The Missing Link in Apicomplexan Mitochondrial Metabolism Is Required for Full Virulence of and
- Helminth Infections, Type-2 Immune Response, and Metabolic Syndrome
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



