-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaHDA6 Regulates Locus-Directed
Heterochromatin Silencing in Cooperation with MET1
Heterochromatin silencing is pivotal for genome stability in eukaryotes. In
Arabidopsis, a plant-specific mechanism called
RNA–directed DNA methylation (RdDM) is involved in heterochromatin
silencing. Histone deacetylase HDA6 has been identified as a component of such
machineries; however, its endogenous targets and the silencing mechanisms have
not been analyzed globally. In this study, we investigated the silencing
mechanism mediated by HDA6. Genome-wide transcript profiling revealed that the
loci silenced by HDA6 carried sequences corresponding to the RDR2-dependent
24-nt siRNAs, however their transcript levels were mostly unaffected in the
rdr2 mutant. Strikingly, we observed significant overlap of
genes silenced by HDA6 to those by the CG DNA methyltransferase MET1.
Furthermore, regardless of dependence on RdDM pathway, HDA6 deficiency resulted
in loss of heterochromatic epigenetic marks and aberrant enrichment for
euchromatic marks at HDA6 direct targets, along with ectopic expression of these
loci. Acetylation levels increased significantly in the hda6
mutant at all of the lysine residues in the H3 and H4 N-tails, except H4K16.
Interestingly, we observed two different CG methylation statuses in the
hda6 mutant. CG methylation was sustained in the
hda6 mutant at some HDA6 target loci that were surrounded
by flanking DNA–methylated regions. In contrast, complete loss of CG
methylation occurred in the hda6 mutant at the HDA6 target loci
that were isolated from flanking DNA methylation. Regardless of CG methylation
status, CHG and CHH methylation were lost and transcriptional derepression
occurred in the hda6 mutant. Furthermore, we show that HDA6
binds only to its target loci, not the flanking methylated DNA, indicating the
profound target specificity of HDA6. We propose that HDA6 regulates
locus-directed heterochromatin silencing in cooperation with MET1, possibly
recruiting MET1 to specific loci, thus forming the foundation of silent
chromatin structure for subsequent non-CG methylation.
Published in the journal: . PLoS Genet 7(4): e32767. doi:10.1371/journal.pgen.1002055
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002055Summary
Heterochromatin silencing is pivotal for genome stability in eukaryotes. In
Arabidopsis, a plant-specific mechanism called
RNA–directed DNA methylation (RdDM) is involved in heterochromatin
silencing. Histone deacetylase HDA6 has been identified as a component of such
machineries; however, its endogenous targets and the silencing mechanisms have
not been analyzed globally. In this study, we investigated the silencing
mechanism mediated by HDA6. Genome-wide transcript profiling revealed that the
loci silenced by HDA6 carried sequences corresponding to the RDR2-dependent
24-nt siRNAs, however their transcript levels were mostly unaffected in the
rdr2 mutant. Strikingly, we observed significant overlap of
genes silenced by HDA6 to those by the CG DNA methyltransferase MET1.
Furthermore, regardless of dependence on RdDM pathway, HDA6 deficiency resulted
in loss of heterochromatic epigenetic marks and aberrant enrichment for
euchromatic marks at HDA6 direct targets, along with ectopic expression of these
loci. Acetylation levels increased significantly in the hda6
mutant at all of the lysine residues in the H3 and H4 N-tails, except H4K16.
Interestingly, we observed two different CG methylation statuses in the
hda6 mutant. CG methylation was sustained in the
hda6 mutant at some HDA6 target loci that were surrounded
by flanking DNA–methylated regions. In contrast, complete loss of CG
methylation occurred in the hda6 mutant at the HDA6 target loci
that were isolated from flanking DNA methylation. Regardless of CG methylation
status, CHG and CHH methylation were lost and transcriptional derepression
occurred in the hda6 mutant. Furthermore, we show that HDA6
binds only to its target loci, not the flanking methylated DNA, indicating the
profound target specificity of HDA6. We propose that HDA6 regulates
locus-directed heterochromatin silencing in cooperation with MET1, possibly
recruiting MET1 to specific loci, thus forming the foundation of silent
chromatin structure for subsequent non-CG methylation.Introduction
Chromatin modification is epigenetic information that has evolved in diverse eukaryotes adding another layer of information to the DNA code. In higher eukaryotes, histone modification and DNA methylation are involved in numerous biological processes such as development, regeneration, and oncogenesis [1], [2]. In addition, the eukaryotic genome has evolved epigenetic mechanisms to silence potentially harmful transposable elements (TEs) and the repetitive elements that constitute a large proportion of the genome [3]. Heterochromatin formation, a striking function of the eukaryotic genome, is intricately controlled through repressive histone modification and DNA methylation [4]. Thus, mutations that affect the status of chromatin structure often result in strong phenotypic alterations or inviability, because of aberrant regulation of gene expression or distorted genome stability [5]–[8].
The flowering plant, Arabidopsis thaliana, is a model organism particularly suited for epigenetic research due to the availability of viable and heritable null mutants of histone modifying enzymes and DNA methyltransferases. Recent genome-wide studies on epigenetic marks of gene silencing in plants have focused on DNA methylation or repressive histone methylations [9]–[13]. Few studies, however, have focused on histone deacetylation, which is crucial for epigenetic regulation in eukaryotes [14], [15]. Investigating histone deacetylation and DNA methylation in Arabidopsis could contribute not only to our understanding of plant biology, but also to a broad range of essential biological processes in mammals and therapeutic applications in humans [16], [17].
Gene silencing has been investigated extensively in Arabidopsis. Plants have evolved gene silencing machinery called RNA-directed DNA methylation (RdDM). Plant-specific RNA POLYMERASE IV (Pol IV), RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) and DICER-LIKE 3 (DCL3) are involved in the production of 24-nt small interfering RNAs (siRNAs) that guide DNA methyltransferases, DOMAINS REARRANGED METHYLTRANSFERASES 1/2 (DRM1/2), to the corresponding genomic DNA for de novo DNA methylation in all cytosine contexts (CG, CHG, CHH; H: A, T, or C; [18]). METHYLTRANSFERASE 1 (MET1), a homolog of mammalian DNMT1, is primarily responsible for the maintenance of genome-wide CG methylation [19]–[22]. KRYPTONITE (KYP), a member of the Su(var)3–9 class of histone methyltransferases, contributes an epigenetic mark of constitutive heterochromatin, histone H3 Lys 9 dimethylation (H3K9me2) [23], [24]. CHROMOMETHYLASE 3 (CMT3), a plant-specific DNA methyltransferase, maintains CHG methylation via H3K9me2 dependence mediated by KYP [23], [25], [26]. Histone Deacetylase 6 (HDA6), a homolog of yeast RPD3 and mammalian HDAC1, is involved in gene silencing and RNA-directed DNA methylation [27]–[30].
Of the 16 Arabidopsis histone deacetylases [31], the importance of HDA6 in gene silencing was discovered by identification of HDA6 in three independent genetic screens of gene silencing [27], [32], [33]. In each case, hda6 mutant plants lacking histone deacetylase activity (sil1, axe1, and rts1) were shown to exhibit reactivation of transcription on target transgenes. Analyses of the endogenous function of HDA6 have been limited, thus far, to the regulation of chromatin at repetitive sequences such as rDNA loci [28], [30], [34], [35], transposable elements and centromeric satellite repeats [29], [36]. However, the positions of the loci silenced by HDA6 have yet to be determined genome-wide.
Various effects of the hda6 mutations on cytosine methylation have been observed previously. Several transposable elements were hypomethylated in sil1 [36]. Reduction of DNA methylation has been reported for the siRNA-directed NOS promoter in rts1, predominantly at CG and CHG sites [27]. Similarly, a reduction in CG and CHG methylation was observed in axe1-5, sil1, and rts1 mutants at rDNA repeats, although the demethylation was much less than that observed in the DNA hypomethylation mutant ddm1 [28], [30]. In contrast to these observations, a drastic reduction in CHG methylation, but not CG methylation, was observed in a Sadhu-type transposable element in axe1-5 [37], and 5S rDNA in sil1 [35]. Furthermore, few changes in DNA methylation were observed in the centromeric repeats or transgene region in sil1, although their silencing was lost [29], [38]. These various effects of the hda6 mutations on DNA methylation might be due to locus dependence rather than differences in the mutations themselves, because similar effects were observed between the mutants [28], [30]. Because previous studies have focused on only a few specific loci, precisely how the hda6 mutation influences DNA methylation in general remains obscure. Therefore, a genome-wide analysis of HDA6 target loci is vital to improve our understanding of the mechanistic basis for HDA6-mediated gene silencing via DNA methylation and histone modification.
In this study, aimed at understanding the silencing mechanism mediated by HDA6, we identified HDA6 transcriptionally repressed loci across the genome and determined the direct targets of HDA6. We also studied the regulation mechanisms involved in histone modification and DNA methylation on HDA6 direct targets. Our data show that the hda6 mutation causes loss of heterochromatic marks and aberrant enrichment for euchromatic epigenetic marks at HDA6 direct targets. Furthermore, we present evidence that the upregulated loci in hda6 overlapped with those in met1, and that the hda6 mutation causes the complete loss of DNA methylation on some HDA6 target loci. These results suggest that a strong functional connection between HDA6 and MET1 exists. Remarkably, hypomethylation only occurred in hda6 on the HDA6 target loci where surrounding MET1 targets were absent. We propose, therefore, that HDA6 is required for gene silencing and that it acts in cooperation with MET1 to build the infrastructure of heterochromatin.
Results
Genome-Wide Identification of Loci Derepressed in axe1-5
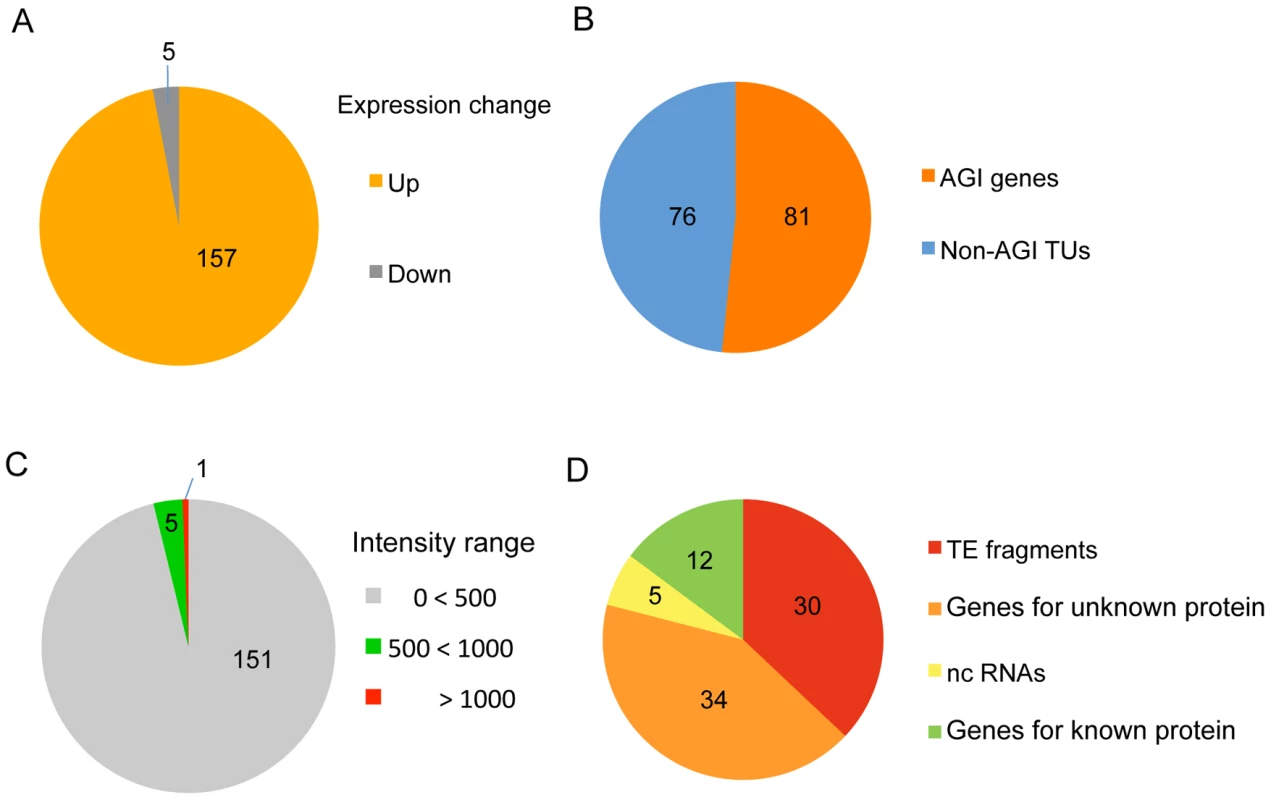
To identify target loci for HDA6 binding in the Arabidopsis genome, we first performed a genome-wide comparison of RNA accumulation between the wild-type plant (DR5) and the hda6 mutant, axe1-5 [33], using a whole-genome tiling array. This approach identified 157 statistically significant loci that were transcriptionally upregulated in axe1-5 compared with wild-type plants (>3 fold, p-initial<10−6, FDR α = 0.05) (Figure 1A and 1B). RT–PCR of a random selection of these loci was used to confirm their up-regulation in the axe1-5 mutant (Figure S1). Among these loci, nearly half (81 genes; see Table S1) were annotated by the Arabidopsis Genome Initiative (http://www.arabidopsis.org/; hereafter referred to as AGI genes). The other half (76 genes; Table S2) were intergenic non-AGI annotated transcriptional units (non-AGI TUs) identified using the ARTADE program [39] (Figure 1B). It is noteworthy that only a small fraction of transcripts (5 loci: 3% of all differentially expressed loci) were classified as having reduced levels of expression in axe1-5 (Figure 1A, Table S3). We consistently found that the loci upregulated in axe1-5 were strongly silenced in wild-type plants (Figure 1C) and consisted predominantly of TE fragments and genes for unknown proteins (79%) (Figure 1D). A survey of the TE fragments [40], mapping on or around the loci upregulated in axe1-5 (from 1 kb upstream to 1 kb downstream), showed that a significant number of the fragments (342 TE fragments) were located on or around such loci (Table S1 and S2). These results show that HDA6 regulates gene silencing on a genome-wide scale.
HDA6-Mediated Gene Silencing Is Mostly Independent of the RdDM Components
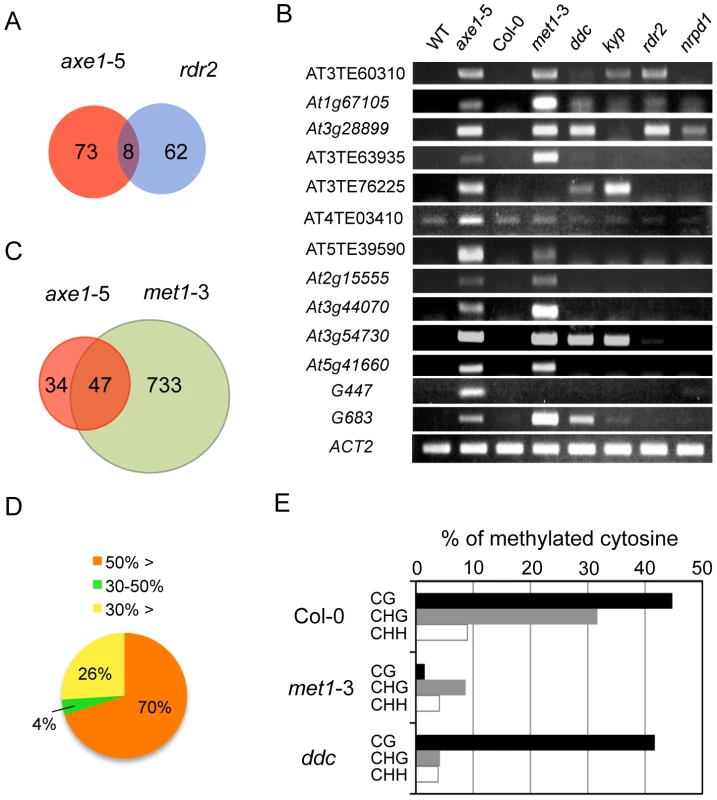
Forward genetic screens for plants deficient in RNA-mediated transcriptional silencing identified HDA6 as an essential component of the RdDM pathway [27], [41]. To address whether the endogenous HDA6 target loci were also directed by the RdDM pathway, siRNAs from the ASRP database [42] were mapped to the loci derepressed in axe1-5. Consistent with knowledge that 24-nt-long siRNAs are required for the establishment of RdDM, the most abundant siRNAs mapping to upregulated loci in axe1-5 are 24-nt long (Figure S2A). These 24-nt siRNAs were hardly found in the rdr2 and dcl3 mutants (Figure S2B), suggesting that loci derepressed in axe1-5 contain siRNA sequences produced by RDR2 and DCL3-dependent pathways, as previously predicted [27], [41]. Thus, our previous study for the targets of RDR2 [43], that were identified using same growth conditions and array technology as in this study, were compared with the genes derepressed in axe1-5. This revealed, surprisingly, despite the loss of 24-nt siRNAs from those genes were observed in the rdr2 mutant (Figure S2B), the majority of the loci derepressed in axe1-5 are kept in a silenced state in the rdr2 mutant (Figure 2A). In fact, elevated transcript levels were not detectable at many loci in the mutants deficient in siRNA production (rdr2, and nrpd1; 10 and 11 respectively, out of 13 genes tested; Figure 2B). There also was evidence that small subsets of the HDA6-mediated gene silencing showed dependence on RdDM pathway and the overlapped genes between axe1-5 and rdr2 was confirmed for their accumulated transcripts (AT3TE60310, At1g67105, and At3g28899) in the RdDM mutants (rdr2 and nrpd1; Figure 2B). Interestingly, larger overlap to the triple mutant drm1 drm2 cmt3 (ddc) involved in siRNA-directed non-CG methylation was observed (5 out of 13 genes; Figure 2B). Taken together, these results indicate the partial involvement of RdDM pathway in HDA6-mediated endogenous gene silencing.
HDA6 and the CG DNA Methyltransferase MET1 Share Common Target Loci for Epigenetic Silencing
We also examined the effects of mutations in other chromatin modifying enzymes on the silencing of putative HDA6 target loci. Strikingly, 10 out of 13 of the putative HDA6 target loci were also upregulated in the met1-3 mutant (Figure 2B). To address whether HDA6 and MET1 share common target loci genome-wide, we also identified differentially regulated loci in met1-3 using a tiling array (Tables S4, S5, S6 and S7), and compared the upregulated loci in met1-3 with those in axe1-5. A significant overlap of upregulated loci in axe1-5 to those in met1-3 was observed (Hypergeometric distribution, P = 1.08E−54; Figure 2C). Furthermore, the DNA methylation status of the loci derepressed in axe1-5 was also investigated using publicly available DNA methylation datasets [13]. Most of the genes upregulated in axe1-5 (i.e. 70% of the upregulated AGI genes) were substantially methylated in the wild-type plants with more than 50% of all cytosines at regions surrounding transcriptional start sites methylated (Figure 2D). Cytosine methylation in the wild-type plants was predominantly found at CG, to a lesser extent at CHG, and least of all at the CHH sites of derepressed AGI genes in axe1-5 (Figure 2E). A large proportion of the cytosine methylation on derepressed AGI genes in axe1-5 appears to be highly dependent on MET1 because the drastic reduction in cytosine methylation was observed not only at CG, but also CHG and CHH sites (Figure 2E). In contrast, CG methylation in the ddc mutant remained at similar levels as the wild-type plants (Figure 2E). Thus, these data demonstrate that the CG DNA methyltransferase MET1 is required for HDA6-mediated epigenetic gene silencing.
Identification of the Direct Targets of HDA6
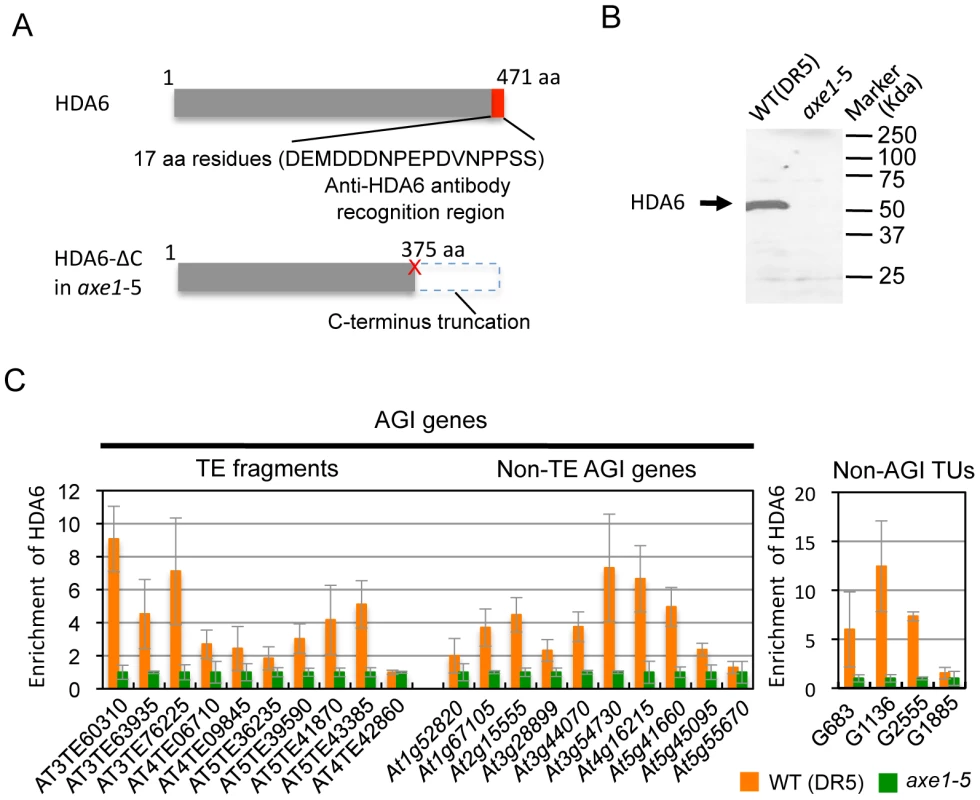
Identification of the direct targets of HDA6 is a crucial step in providing mechanistic insight into HDA6 function in transcriptional control, chromatin regulation, and DNA methylation. To determine the HDA6 target loci using chromatin immunoprecipitation (ChIP), we raised a specific antibody against HDA6. The epitope for the antibody was designed against the C-terminal region of the HDA6 protein that is absent in axe1-5. We verified that the peptide sequence was not similar to any other sequence of annotated Arabidopsis proteins. Thus, comparisons made between wild-type and axe1-5 for the level of enrichment of immunoprecipitated DNA using the antibody allowed us to exclude any non-specific signals to identify HDA6 binding sites (Figure 3A). Western blot analysis confirmed the specificity of the antibody, which detected a unique band of 53 kDa in the wild-type, but not in axe1-5 (Figure 3B).
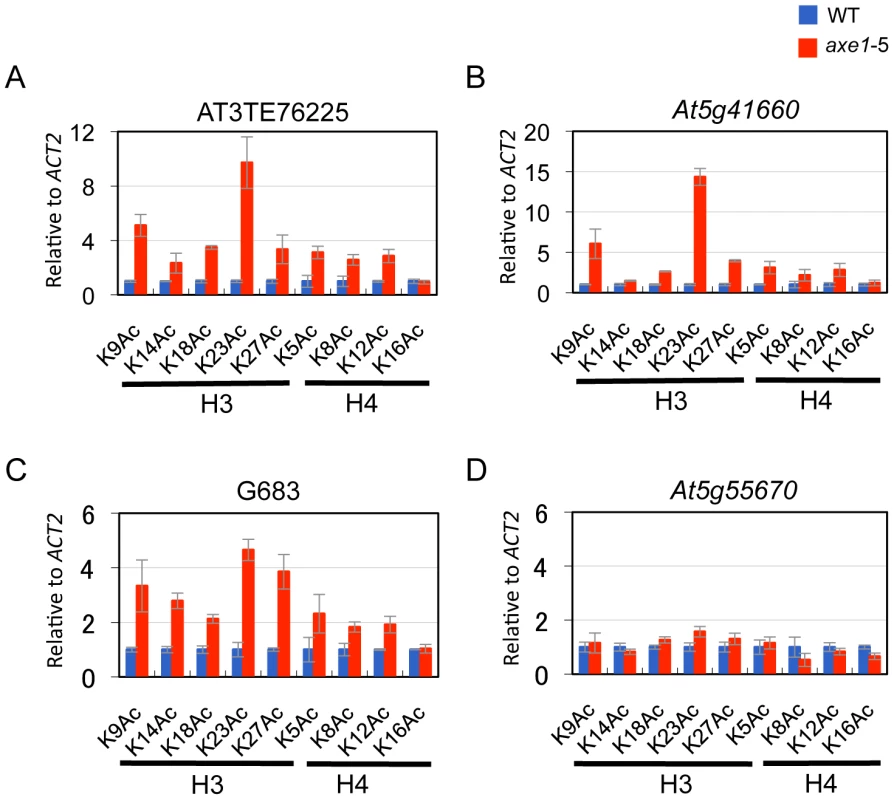
Using the HDA6 antibody, we performed ChIP assays and quantitative PCR (qPCR). Three genes, AT3TE60310 (At3g42658), AT3TE76225 (At3g50625) and At5g41660 were selected from genes upregulated in axe1-5, representing RdDM dependent, MET1 independent, and MET1 dependent genes, respectively (Figure S1). Three primer sets were designed for each gene within the promoter, 5′ and 3′ regions of the genes (Figure S3). HDA6 binding levels in the wild-type plants were significantly higher than in axe1-5 for all of the genes tested, regardless of the dependence on RdDM pathway or MET1. This indicates that HDA6 binds directly to all such genes. In addition, preferential binding of HDA6 was observed within the 5′ regions of the genes. Therefore, a further screening of HDA6 target loci was performed for the 5′ regions of selected loci from those upregulated in axe1-5 (Figure S1; Figure 3C). As a negative control, we tested one gene that exhibited no apparent transcriptional change (At5g55670, Figure S1), and found only a minimal difference. Our experiments show that HDA6 binding levels were enriched by 2 to 12-fold in the wild-type plants relative to axe1-5, with statistical significance observed for 17 of the loci (Figure 3C). 5 loci did not show significant differences between the wild-type and axe1-5 mutant.
HDA6 Is Required for Heterochromatic Silencing, and the Mutation Results in Loss of Heterochromatic Histone Modification Along with Aberrant Enrichment for Euchromatic Modification at HDA6 Targets
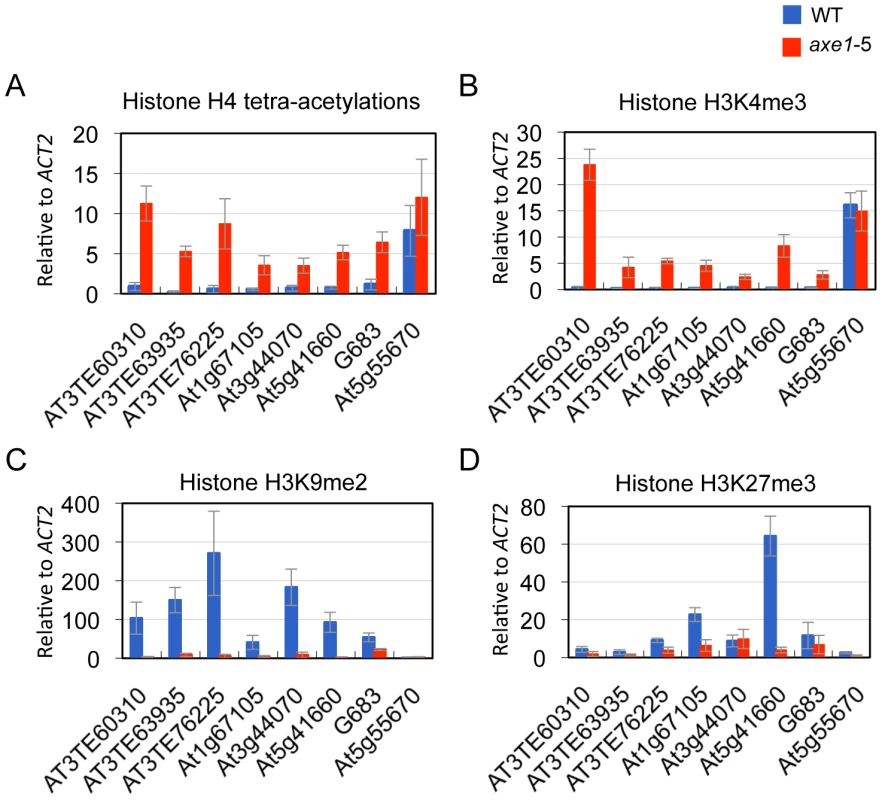
Heterochromatic or repressive regions are associated with H3K9me2 and/or H3K27me3, whereas euchromatic or transcriptionally active regions are associated with H3K4me3 and H4 tetra-acetylation (H4 tetra-acetylated on K5, K8, K12, and K16) [44]. Furthermore, many endogenous RdDM targets are known to be associated with euchromatic modification H3K4me3 [45]. To elucidate HDA6 function and chromatin status, the effects of the hda6 mutation on histone modification was analyzed using ChIP-qPCR on its direct targets. The results show that, regardless of the dependence on siRNAs or positions on the chromosome, weak H4 tetra-acetylation and H3K4me3 and significantly high levels of heterochromatic modification, H3K9me2, were observed in the wild-type plants at all of the loci tested (Figure 4A, 4B and 4C; Figure S4). In the axe1-5 mutant, the active marks strongly increased (H4 tetra-acetylation at range 5 to 30 fold and H3K4me3 at 8 to 78 fold, respectively), and the levels of H3K9me2 were drastically reduced (range 2 to 53 fold) compared with the wild-type plants (Figure 4A, 4B and 4C; Figure S4). H3K27me3, another repressive modification, are highly enriched in wild-type plants predominantly on genes within the chromosome arm regions, such as At1g67105 and At5g41660, and drastically reduced in axe1-5 (Figure 4D). Consistent with this observation, the enrichment of H3K27me3 in the euchromatic arm regions was also seen in the previous genome-wide studies of H3K27me3 [11], [12]. These results indicate that the hda6 mutation caused an alteration of the chromatin status from a heterochromatic to euchromatic state that was concomitant with transcriptional release of HDA6 target loci.
HDA6 deacetylase activity has been reported for H3K9, H3K14, H4K5 and H4K12, as well as for H4 tetra-acetylation [34]. The activity at other residues, however, is currently unknown. To assess the HDA6 deacetylase activity at other lysine residues, we investigated all of the potential acetylation sites of the H3 and H4 N-tails, including pre-determined sites using ChIP-qPCR. Three HDA6 target loci (AT3TE76225, At5g41660, and G683) were examined. The results showed that in axe1-5, the acetylation levels significantly increased at H3K9, H3K14, H3K18, H3K23, H3K27, H4K5, H4K8, and H4K12 residues relative to those in wild-type plants at all three loci tested (Figure 5A, 5B and 5C, Figure S5). Interestingly, among these residues, H3K23ac levels showed the highest enrichments in axe1-5, for the three loci, AT3TE76225, At5g41660 and G683 (at 10, 14, 5 fold respectively). The acetylation levels were not significantly altered in axe1-5 for the control genes At5g55670 (Figure 5D) and ACT2 (Figure S5). It is noteworthy that the deacetylase activity observed for HDA6 at H3K27ac as well as H3K9ac, are both likely to be important for the subsequent histone methylation of H3K27me3 and H3K9me2, respectively [46]. Interestingly, residues that showed increased levels of acetylation in axe1-5, were identical to the target residues in yeast RPD3 deacetylation [47]. These results indicate that the deacetylase activity of HDA6 occurred on all of the lysine residues in H3 and H4 N-tails, except H4K16.
The Effect of the hda6 Mutation on DNA Methylation
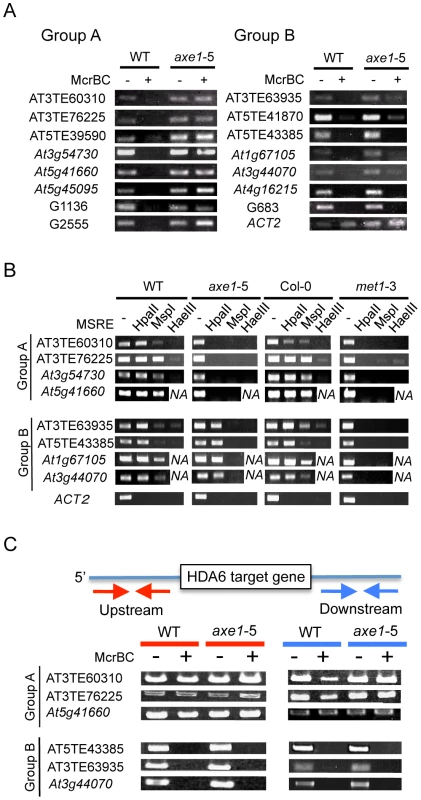
The different effects that hda6 mutations impose on DNA methylation have been reported as described above. To assess the function of HDA6 on DNA methylation and the relationship between HDA6 and MET1, the DNA methylation status of HDA6 direct targets was investigated. We used the endonuclease McrBC (Figure 6A), which preferentially cleaves methylated DNA, and a Chop-PCR assay using methylation sensitive restriction enzymes (Figure 6B), whose cleavage is blocked by DNA methylation.
In the McrBC assay shown in Figure 6A, no strong bands were detected in the wild-type plants after McrBC digestion, indicating that the direct targets of HDA6 are highly DNA methylated in the wild-type plants. This is consistent with dense DNA methylation on the loci upregulated in axe1-5 (Figure 2D and 2E). However, in axe1-5, we observed two types of McrBC sensitivity among the HDA6 direct target loci. The Group A genes substantially lost cytosine methylation, as demonstrated by the presence of strong bands of similar intensity in both Group A genes and the non-digested control (Figure 6A, left panel). In contrast, the genes in Group B retained DNA methylation in axe1-5 in common with the wild-type plants as no strong bands were detected (Figure 6A, right panel).
We also performed Chop-PCR assays to investigate in which cytosine contexts are dependent on HDA6 (Figure 6B). The methylation sensitive restriction enzymes used were HpaII, MspI and HaeIII, which reports CG, CHG, and CHH methylation, respectively. From each group categorized in Figure 6A, four representative HDA6 target loci were tested and using ACT2 as a negative control. PCR amplification of ACT2 was undetectable regardless of the genotypes or the enzymes tested (Figure 6B), confirming substantial cleavage by the enzymes had occurred. In the wild-type plants, strong amplification, at a similar level as the undigested control was detected after digestion with HpaII (CG methylation). Strong amplification was observed with MspI (CHG methylation), but it was mostly less than the undigested control; least amplification of all was observed with HaeIII (CHH methylation) (Figure 6B). The results of this experiment are consistent with the results shown in Figure 2E and Figure 6A, where HDA6 target loci were shown to be significantly methylated in wild-type plants, predominantly at CG sites, and to a similar or lesser extent at CHG sites, but least of all at CHH sites.
In common with the results obtained for the McrBC assay (Figure 6A); the results from the Chop-PCR assays split the HDA6 target loci into two separate groups. In Group A (AT3TE60310, AT3TE76225, At3g54730 and At5g41660), complete demethylation in axe1-5 occurred in all sequence contexts and amplification after digestion of each enzyme was undetectable (Figure 6B, Group A). In the Group B genes (AT3TE63935, AT5TE43385, At1g67105, and At3g44070), for axe1-5, however, CG methylation was mostly sustained at the similar level as the wild-type (Figure 6B, lower panel, HpaII digest), although a drastic reduction in CHG and CHH methylation was detected [Figure 6B, lower panel CHG (MspI) and CHH (HaeIII)]. Bisulfite sequencing analyses of wild-type, axe1-5 and met1-3 further confirmed that Group A genes (At5g41660 and AT3TE76225) lost DNA methylation in all sequence contexts in axe1-5, and a Group B gene (AT3TE63935) lost CHG and CHH methylation but sustained CG methylation in axe1-5 (Figure S6). Collectively, the methylation analyses of the HDA6 targets demonstrated that, 1) CG and CHG sites were predominantly highly methylated in the wild-type plants; 2) CHG and CHH methylation was substantially reduced at all target loci in axe1-5; and, 3) CG methylation was lost at some loci but sustained at others in axe1-5.
An Absence of DNA Methylated Regions around HDA6 Target Loci Is Correlated with the Loss of CG Methylation in axe1-5
Why were two different CG methylation states observed in axe1-5? Interestingly, according to the public database for DNA methylation [9], a clear correlation was observed between a loss of CG methylation on the HDA6 targets in axe1-5 and the absence of methylated DNA regions around the HDA6 target loci (Figure S7). HDA6 target loci in Group A (with loss of DNA methylation) were shown to be isolated from other methylated DNA regions, whereas HDA6 target loci in Group B (with persistent CG methylation) were surrounded by other methylated DNA regions. We confirmed the presence or absence of DNA methylated regions around HDA6 target loci (Figure 6C). The 1.5 kb regions upstream and downstream of the HDA6 target loci were analyzed to determine their DNA methylation status using McrBC assays. We found evidence of robust DNA methylation around the HDA6 target loci in Group B (with persistent CG methylation in axe1-5) (Figure 6C, Group B). These methylated regions often contained other TE fragments adjacent to HDA6 target loci. These TEs were densely DNA methylated dependently on MET1 but independently of HDA6, since the PCR amplification after McrBC digestion was detected only in met1-3 (Figure S8). We also confirmed that these adjacent TE fragments were not targeted by HDA6 using ChIP-qPCR assays; these results showed no enrichment of HDA6 binding to adjacent TE fragments in the wild-type plants compared with axe1-5 (Figure S9). As a result, HDA6 target loci with sustained CG methylation in axe1-5 (Group B) must harbor the flanking TE fragments that are highly DNA methylated by MET1 independently of HDA6. On the other hand, we found that the target loci in Group A (with loss of DNA methylation in axe1-5) were isolated from other DNA methylated regions as no substantial methylation was detected around the target loci (Figure 6C, Group A). We confirmed the absence of DNA methylation around the HDA6 target loci in Group A by bisulfite sequencing analysis of the upstream region of a Group A gene, At5g41660 (data not shown). Thus, we deduced a requirement for HDA6 involvement in CG methylation by MET1 for HDA6 target loci, in the absence of other flanking DNA methylated regions.
Discussion
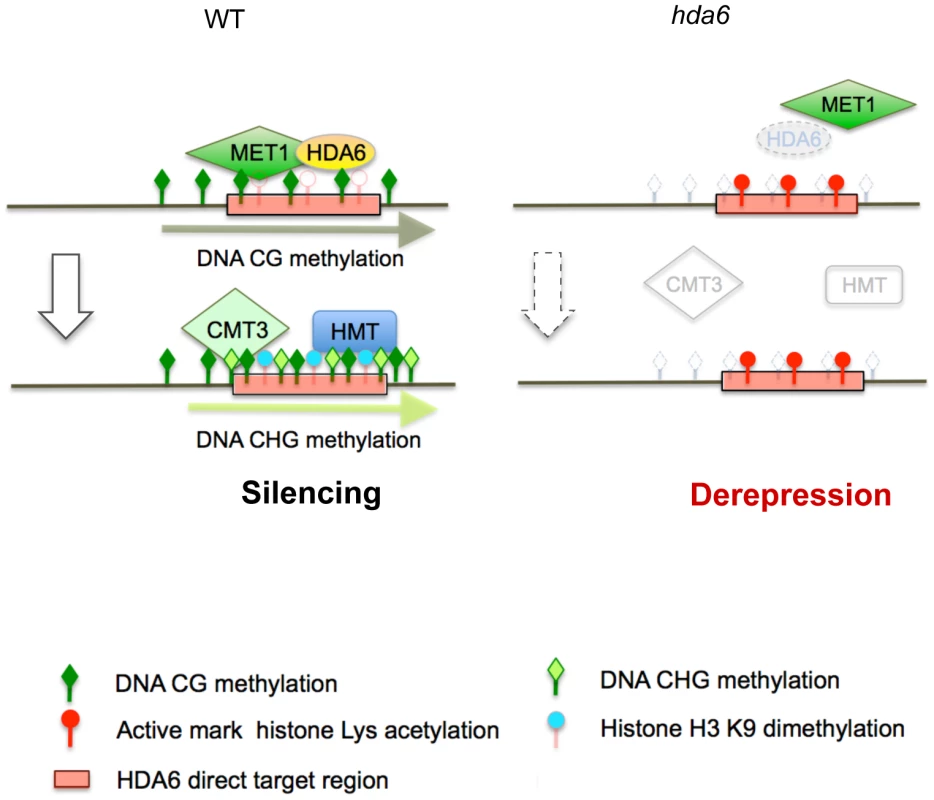
We have identified 157 loci that require HDA6 for epigenetic silencing in Arabidopsis. This is the first report to identify derepressed loci in axe1-5 on a genome-wide scale. Our study revealed several interesting features of HDA6 target loci, mapped large numbers of TE fragments and DNA methylation sites in wild-type Arabidopsis plants. The derepressed loci in axe1-5 overlapped significantly with the derepressed loci in met1-3, a CG DNA methyltransferase mutant, rather than the RdDM deficient mutants rdr2 and ddc, suggesting that HDA6 plays an important role in gene silencing, in cooperation with MET1. We also identified 17 direct targets of HDA6 using ChIP-qPCR assays with a HDA6 specific antibody. We found that HDA6 was required for heterochromatic histone modifications and DNA methylation in the target loci. Interestingly, HDA6 deficiency resulted in aberrant enrichment for euchromatic epigenetic marks and DNA hypomethylation at HDA6 targets, along with ectopic expression of these loci. DNA hypomethylation at CG sites in axe1-5 occurred at some HDA6 target loci, but only where isolated from other MET1 target loci, possibly indicating the requirement of HDA6 for the recruitment of MET1 to specific loci.
We showed that all of the HDA6 direct targets tested in this study colocalized with a constitutive heterochromatin mark, histone H3K9me2 (7 out of 7; Figure 4C), rather than another mark of repressive chromatin, H3K27me3 (Figure 4D, 3 out of 7). We saw no evidence of euchromatic modification H3K4me3 (0 out of 7; Figure 4B). Moreover, our results suggested the deacetylase activity of HDA6 against H3K9ac and H3K27ac, will be important for subsequent histone methylations of H3K9me2 and H3K27me3, as well as H3K14ac, H3K18ac, H3K23ac, H4K5ac, H4K8ac and H4K12ac. These results suggest that H3K9 and H3K27 deacetylation by HDA6 is essential for the establishment of the heterochromatic and repressive marks mediated by H3K9me2 and H3K27me3, and HDA6 deficiency resulted in loss of heterochromatic histone marks and aberrant enrichment for euchromatic marks at HDA6 target loci. It is noteworthy that the enrichments observed on H3K23ac were the highest among the possible acetylation sites, which may indicate the importance of H3K23 deacetylation on heterochromatic gene silencing. The abundance of silenced TE fragments and genes for unknown proteins among the loci derepressed in axe1-5 (Figure 1C and 1D) supports the role of HDA6 in silencing at heterochromatic regions. In addition, an important role for HDA6 in CHG methylation is indicated by the observation that all of the direct targets of HDA6 tested in this study lost CHG methylation in axe1-5 (Figure 6B; Figure S6). Several papers report that CHG methylation maintained by CMT3 is dependent on H3K9me2 and that CMT3 is recruited to methylated histones [24]–[26]. Taken together, this indicates that HDA6 deacetylase activity against its target loci is required for establishment of the heterochromatic and repressive marks H3K9me2 and H3K27me3, and CHG methylation by CMT3.
The separation of endogenous HDA6 target loci from endogenous RdDM target loci were investigated in this study. Our results show that endogenous HDA6 target loci were associated with a constitutive heterochromatic mark, H3K9me2 (Figure 4C), but not a euchromatic mark, H3K4me3 (Figure 4B). However, many of the endogenous target loci of RdDM components (such as Pol V and DRD1) were found to be associated with euchromatic histone modification H3K4me3, but not H3K9me2 [45]. Surprisingly, genome-wide identification of the loci derepressed in axe1-5 revealed that these loci overlap with only a small fraction of the genes upregulated in rdr2 (Figure 2A). However, considering the recent studies proposing the role of siRNAs in re-establishment of DNA methylation and gene silencing when DNA methylation was lost in the DNA methylation deficient mutants like met1 and ddm1 [48]–[50], the siRNAs found on the HDA6 target loci might also have a role in this mechanism, and therefore the double mutants of hda6 and siRNA deficient mutants might result in larger release of gene silencing. In addition, cell-type specific regulation of siRNAs and TEs silencing especially in the gametes has been proposed recently [51]. In this case, DDM1 expression is downregulated in the pollen vegetative nucleus, which accompanies the sperm cells. Considering that HDA6 has common features with DDM1 [18], [29], [36]–[38], it would also be interesting to see if HDA6 also have a role in regulating transposon silencing in gametes.
An important functional connection between HDA6 and MET1 was revealed by observing the significant overlap of the loci upregulated in axe1-5, with the loci upregulated in met1-3 (Figure 2B and 2C). Indeed, we found several HDA6 target loci (AT3TE60310, AT3TE76225, At3g54730 and At5g41660) that require HDA6 for MET1 CG methylation (Figure 6B; Group A). Thus, we propose that HDA6 acts in cooperation with MET1, possibly as a recruiter or as a component of the silencing machinery with MET1 (Figure 7). Actually we observed the loss of HDA6 binding on several HDA6 targets in the met1-3 mutant (Figure S10). It indicates the requirement for MET1 and/or CG methylation to facilitate HDA6 binding, suggesting the cooperative interplay between HDA6 and MET1. Further support for this hypothesis, is the observation that HDA6 targets isolated from other flanking MET1 targets experienced a loss of CG methylation in the absence of HDA6 (Figure 6B and 6C). It is also supported by several papers evidencing the physical interactions that occur between histone deacetylases and DNA methyltransferases in mammals [52]–[54].
The sustained CG methylation status of the other HDA6 targets in the axe1-5 mutant was found to correlate with the existence of other flanking MET1 target loci in neighboring regions of the HDA6 targets (Figure 6B and 6C, Group B; Figure S7, S8, S9, S11). Thus it appears likely, therefore, that MET1 could be recruited to the neighboring regions of the HDA6 targets and pass through the HDA6 targets (Figure S11). Another possibility is that MET1-dependent CG methylation is the primary repressive modification to silence those HDA6 targets. There also were many loci that were derepressed only in the met1-3 mutant (Figure 2C, Figure S12).
It is noteworthy that the intensities of the RT-PCR bands were greater in met1-3 than in axe1-5 for the genes with sustained CG methylation (i.e. At1g67105, AT3TE63935, At2g15555, At3g44070, and G683; Figure 2B), indicating that sustained CG methylation on the HDA6 target loci can be repressive to some extent. However, there is clear evidence that these genes are transcriptionally derepressed strongly in the absence of HDA6 or MET1, regardless of the CG methylation status. We deduce, therefore, that both HDA6 histone deacetylation and MET1 CG methylation are essential for the silencing of HDA6 target loci. In addition, for most of the HDA6 target loci, CHG and CHH methylation was lost in both hda6 and met1 mutants (Figure 6B). A drastic reduction in CHG and CHH methylation was also observed for the loci silenced by HDA6 in the met1 mutant (Figure 2E). These observations indicate that CHG and CHH methylation on HDA6 targets require the presence of epigenetically silent chromatin associated with both histone deacetylation and CG methylation. Because HDA6 was identified as RTS1 (RNA-mediated transcriptional silencing 1) and MET1 as RTS2 in RdDM screens [22], [27], we strongly suggest that these two genes are deeply connected to each other, as proposed previously [22], [36]. These insights have an important evolutionary implication; that the histone deacetylase superfamily, one of the most ancient enzymes in eukaryotes [16], may build the foundations for gene silencing and concomitant CG DNA methylation. CG DNA methylation is a conserved modification in higher eukaryotes, quite distinct from the siRNA derived CHG or CHH methylation found only in plants [55], [56].
It is noteworthy that HDA6 has high specificity for target loci within the genome. Moreover, we found that HDA6 binds only to its target loci, not the flanking TE fragments (Figure S9). Relatively low numbers of loci were derepressed in axe1-5 (157 loci), contrasting with the met1-3 mutant, where a total of 1215 loci were derepressed. We consistently detected an insignificant difference in the amount of total methylated DNA in axe1-5, whereas a severe reduction was detected in the met1-3 mutant compared with wild-type plants (Figure S13). It was also reported that the total amount of histone H4 tetra-acetylation or H3K4me3 did not change between axe1-5 and the wild-type plants [28]. From this, we conclude that the target specificity of HDA6 is likely to be precisely controlled, with the effect of the hda6 mutation manifesting itself only in local areas. Furthermore, HDA6, but not MET1, has the ability to trigger de novo chromatin silencing, because only backcrossed hda6/wild-type plants were able to restore DNA methylation, low H3K4me3 levels and a silent transcriptional state, comparable with wild-type plants, unlike met1/wt [36], [37]. These findings, taken together, indicate that HDA6 is a regulator of locus-directed heterochromatin silencing in cooperation with MET1, where it acts possibly as a recruiter or as a component of the chromatin silencing machinery with MET1, thus establishing the foundations for silent chromatin status for the subsequent heterochromatin mark H3K9me2 and non-CG methylation (Figure 7).
Because MET1 is the primary CG DNA methyltransferase encoded in Arabidopsis, regulation of the proper and specific distribution of MET1 CG methylation by employment of HDA6 and/or other possible factors would be an efficient way for the Arabidopsis genome to adapt to several developmental and environmental effects. It will be interesting to see if HDA6 and MET1 form a complex as is the case in mammals [52]–[54], or what information is recognized by these factors to trigger the sequential silencing mechanism. In this study, we identified dozens of loci transcriptionally silenced by HDA6 and several loci directly targeted by HDA6. These results will undoubtedly contribute to an understanding of the complex interplay between histone deacetylation and DNA methylation, revealing mechanistic insights into heterochromatin silencing in higher eukaryotes.
Materials and Methods
Plants and Growth Conditions
Seeds were surface-sterilized and stratified for 4 days at 4°C in the dark. The seeds were then grown in tissue culture plates on MS agar (0.8%) medium supplemented with 1% sucrose under 16 h light/8 h dark for 15 days at 22°C. All experiments used axe1-5 [33], met1-3 [21], ddc (drm1-2 drm2-2 cmt3-11, [57]), kyp (SALK_069326 [58]), rdr2-1 (SAIL_1277_H08 [42]), nrpd1a-3 (SALK_128428 [59]) mutants or DR5 [33] and Col-0 wild-type plants. All plants were the Columbia ecotype.
Whole-Genome Tiling Array Analysis
The GeneChip Arabidopsis tiling array set (1.0F Array and 1.0R Array, Affymetrix) was used. Total RNA was extracted using Isogen reagent (Nippon Gene). Probe synthesis, hybridization, detection, data evaluation with U-test (FDR α = 0.05) and P initial value (P<10−6) was conducted essentially as described previously [60]. Three independent biological replicates were performed for each strand array. Detection of intergenic transcribed units was performed as described previously [39], [60] based on the TAIR8 annotation. A threefold increase or decrease in RNA accumulation was taken as additional criteria for defining the loci upregulated or downregulated in the axe1-5 and met1-3 mutants. Tiling array data are available at the GEO website under the accession number GSE23950.
RT-PCR Analysis
Total RNA was extracted using Plant RNA Purification Reagent (Invitrogen) and subjected to cDNA synthesis using the QuantiTect Reverse Transcription Kit (Qiagen), according to the manufacturer's instructions. The PCR conditions were as follows; pre-incubation for 5 min at 94°C, 30 cycles at 94°C for 30 sec, 58°C for 20 sec, 72°C for 40 sec and a final extension at 72°C for 4 min. Primers are listed in Table S8. The amplified DNA was visualized on a 2% agarose gel stained with ethidium bromide.
Generation of the HDA6 Antibody and Western Blot Analysis
Antibodies against HDA6 were generated as follows; a peptide (DEMDDDNPEPDVNPPSS) corresponding to the C-terminus of HDA6 was synthesized, HPLC purified, conjugated to Bovine Serum Albumin (BSA) and used to immunize two rabbits (Scrum). The antiserum obtained was affinity-purified and used for ELISA and western blot analysis. Total protein extraction was performed on 15-day-old seedlings. Seedlings were ground in liquid nitrogen, suspended in PBS supplemented with 1 mM PMSF, centrifuged, and the supernatant used as a total protein extract. The protein concentration was analyzed using the BioRad Bradford reagent and 50 ug of protein was used for western blot analysis. Western blots prepared by the iBlot Dry Blotting system (Invitrogen) were blocked and incubated with the HDA6 antibody diluted at 1∶500, washed, and incubated with anti-rabbit IgG HRP-conjugated antibodies (GE Healthcare) diluted 1∶5000. The results were visualized using ECL Plus Western Blotting Detection Reagents (GE Healthcare).
Chromatin Immunoprecipitation
ChIP assays were performed essentially as described previously [61]. The antibodies used in this study were: anti-H3K4me3 and H3K9me2 [62]; anti-H3K9ac (ab4441) and H3K14ac (ab1191) from Abcam; anti-H4 tetra-acetylation (06-866), H3K27me3 (07-449), H3K18ac (07-328), H3K23ac (07-355), H3K27ac (07-360), H4K5ac (07-327), H4K8ac (07-328), and H4K12ac (07-595) from Millipore, and H4K16ac (CB-SC-8662-R) from Santa Cruz. The precipitates were analyzed with quantitative PCR (Power SYBR real time reagent and ABI Prism 7000, Applied Biosystems) and the relative amount of each modification was estimated as described previously [63]. Statistical significance of the wild-type plants compared with axe1-5 was determined by Kruskal–Wallis test (P<0.05). The primers used are listed in Table S8.
DNA Methylation Analysis
Genomic DNA was extracted using a Phytopure DNA extraction kit (GE Healthcare) and 5 µg of genomic DNA was linearlized with 20 U BamHI for 3 hours at 37°C. McrBC assays were performed by incubating 30 U of McrBC per 1 µg of BamHI digested genomic DNA at 37°C for 16 hours before PCR amplification as described for RT-PCR with a 1 min extension time. Chop-PCR assays [30] were performed using the methylation sensitive restriction enzymes HpaII, MspI, and HaeIII (NEB). Linearlized genomic DNA was incubated with the enzymes (30 U/µg) at 37°C for 3 hours and subjected to PCR analysis. The amplified DNA was visualized on a 1.0% agarose gel stained with ethidium bromide.
Supporting Information
Zdroje
1. HendersonIRJacobsenSE
2007
Epigenetic inheritance in plants.
Nature
447
418
424
2. HoLCrabtreeGR
2010
Chromatin remodelling during development.
Nature
463
474
484
3. SlotkinRKMartienssenR
2007
Transposable elements and the epigenetic regulation of the
genome.
Nat Rev Genet
8
272
285
4. CedarHBergmanY
2009
Linking DNA methylation and histone modification: patterns and
paradigms.
Nat Rev Genet
10
295
304
5. OkanoMBellDWHaberDALiE
1999
DNA methyltransferases Dnmt3a and Dnmt3b are essential for de
novo methylation and mammalian development.
Cell
99
247
257
6. PetersAHO'CarrollDScherthanHMechtlerKSauerS
2001
Loss of the Suv39h histone methyltransferases impairs mammalian
heterochromatin and genome stability.
Cell
107
323
337
7. LaggerGO'CarrollDRemboldMKhierHTischlerJ
2002
Essential function of histone deacetylase 1 in proliferation
control and CDK inhibitor repression.
EMBO J
21
2672
2681
8. MontgomeryRLDavisCAPotthoffMJHaberlandMFielitzJ
2007
Histone deacetylases 1 and 2 redundantly regulate cardiac
morphogenesis, growth, and contractility.
Genes Dev
21
1790
1802
9. ZhangXYazakiJSundaresanACokusSChanSW
2006
Genome-wide high-resolution mapping and functional analysis of
DNA methylation in Arabidopsis.
Cell
126
1189
1201
10. ZilbermanDGehringMTranRKBallingerTHenikoffS
2006
Genome-wide analysis of Arabidopsis thaliana DNA
methylation uncovers an interdependence between methylation and
transcription.
Nat Genet
39
61
69
11. TurckFRoudierFFarronaSMartin-MagnietteMLGuillaumeE
2007
Arabidopsis TFL2/LHP1 specifically associates
with genes marked by trimethylation of histone H3 lysine 27.
PLoS Genet
3
e86
doi:10.1371/journal.pgen.0030086
12. ZhangXClarenzOCokusSBernatavichuteYVPellegriniM
2007
Whole-genome analysis of histone H3 lysine 27 trimethylation in
Arabidopsis.
PLoS Biol
5
e129
doi:10.1371/journal.pbio.0050129
13. ListerRO'MalleyRCTonti-FilippiniJGregoryBDBerryCC
2008
Highly integrated single-base resolution maps of the epigenome in
Arabidopsis.
Cell
133
523
536
14. MillarCBGrunsteinM
2006
Genome-wide patterns of histone modifications in
yeast.
Nat Rev Mol Cell Biol
7
657
666
15. YangXJSetoE
2008
The Rpd3/Hda1 family of lysine deacetylases: from bacteria and
yeast to mice and men.
Nat Rev Mol Cell Biol
9
206
218
16. MinucciSPelicciPG
2006
Histone deacetylase inhibitors and the promise of epigenetic (and
more) treatments for cancer.
Nat Rev Cancer
6
38
51
17. KazantsevAGThompsonLM
2008
Therapeutic application of histone deacetylase inhibitors for
central nervous system disorders.
Nat Rev Drug Discovery
7
854
868
18. MatzkeMABirchlerJA
2005
RNAi-mediated pathways in the nucleus.
Nat Rev Genet
6
24
35
19. FinneganEJDennisES
1993
Isolation and identification by sequence homology of a putative
cytosine methyltransferase from Arabidopsis
thaliana.
Nucleic Acids Res
21
2383
2388
20. VongsAKakutaniTMartienssenRARichardsEJ
1993
Arabidopsis thaliana DNA methylation mutants.
Science
260
1926
1928
21. SazeHMittelsten ScheidOPaszkowskiJ
2003
Maintenance of CpG methylation is essential for epigenetic
inheritance during plant gametogenesis.
Nat Genet
34
65
69
22. AufsatzWMetteMFMatzkeAJMatzkeM
2004
The role of MET1 in RNA-directed de novo and maintenance
methylation of CG dinucleotides.
Plant Mol Biol
54
793
804
23. JacksonJPLindrothAMCaoXJacobsenSE
2002
Control of CpNpG DNA methylation by the KRYPTONITE histone H3
methyltransferase.
Nature
416
556
560
24. JasencakovaZSoppeWJMeisterAGernandDTurnerBM
2003
Histone modifications in Arabidopsis - high
methylation of H3 lysine 9 is dispensable for constitutive
heterochromatin.
Plant J
33
471
480
25. JohnsonLMCaoXJacobsenSE
2002
Interplay between Two Epigenetic Marks: DNA Methylation and
Histone H3 Lysine 9 Methylation.
Curr Biol
12
1360
1367
26. LindrothAMShultismDJasencakovaZFuchsJJohnsonL
2004
Dual histone H3 methylation marks at lysines 9 and 27 required
for interaction with CHROMOMETHYLASE3.
EMBO J
23
4286
4296
27. AufsatzWMetteMFvan der WindenJMatzkeMMatzkeAJ
2002
HDA6, a putative histone deacetylase needed to enhance DNA
methylation induced by double-stranded RNA.
EMBO J
21
6832
6841
28. ProbstAVFagardMProuxFMourrainPBoutetS
2004
Arabidopsis histone deacetylase HDA6 is required
for maintenance of transcriptional gene silencing and determines nuclear
organization of rDNA repeats.
Plant Cell
16
1021
1034
29. MayBPLippmanZBFangYSpectorDLMartienssenRA
2005
Differential regulation of strand-specific transcripts from
Arabidopsis centromeric satellite
repeats.
PLoS Genet
1
e79
doi:10.1371/journal.pgen.0010079
30. EarleyKWPontvianneFWierzbickiATBlevinsTTuckerS
2010
Mechanisms of HDA6-mediated rRNA gene silencing: suppression of
intergenic Pol II transcription and differential effects on maintenance
versus siRNA-directed cytosine methylation.
Genes Dev
24
1119
1132
31. PandeyRMullerANapoliCASelingerDAPikaardCS
2002
Analysis of histone acetyltransferase and histone deacetylase
families of Arabidopsis thaliana suggests functional
diversification of chromatin modification among multicellular
eukaryotes.
Nucleic Acids Res
30
5036
5055
32. FurnerIJSheikhMACollettCE
1998
Gene silencing and homology-dependent gene silencing in
Arabidopsis: genetic modifiers and DNA
methylation.
Genetics
149
651
662
33. MurfettJWangXJHagenGGuilfoyleTJ
2001
Identification of Arabidopsis histone
deacetylase HDA6 mutants that affect transgene expression.
Plant Cell
13
1047
1061
34. EarleyKLawrenceRJPontesOReutherREncisoAJ
2006
Erasure of histone acetylation by Arabidopsis
HDA6 mediates large-scale gene silencing in nucleolar
dominance.
Genes Dev
20
1283
1293
35. VaillantITutoisSCuvillierCSchubertITourmenteS
2007
Regulation of Arabidopsis thaliana 5S rRNA
genes.
Plant Cell Physiol
48
745
752
36. LippmanZMayBYordanCSingerTMartienssenR
2003
Distinct mechanisms determine transposon inheritance and
methylation via small interfering RNA and histone
modification.
PLoS Biol
1
e67
doi:10.1371/journal.pcbi.0010067
37. RangwalaSHRichardsEJ
2007
Differential epigenetic regulation within an
Arabidopsis retroposon family.
Genetics
176
151
160
38. ElmayanTProuxFVaucheretH
2005
Arabidopsis RPA2: A Genetic Link among
Transcriptional Gene Silencing, DNA Repair, and DNA
Replication.
Curr Biol
15
1919
1925
39. ToyodaTShinozakiK
2005
Tiling array-driven elucidation of transcriptional structures
based on maximum-likelihood and Markov models.
Plant J
43
611
621
40. BuisineNQuesnevilleHColotV
2008
Improved detection and annotation of transposable elements in
sequenced genomes using multiple reference sequence sets.
Genomics
91
467
475
41. AufsatzWStoiberTRakicBNaumannK
2007
Arabidopsis histone deacetylase 6: a green link
to RNA silencing.
Oncogene
26
5477
5488
42. KasschauKDFahlgrenNChapmanEJSullivanCMCumbieJS
2007
Genome-wide profiling and analysis of
Arabidopsis siRNAs.
PLoS Biol
5
e57
doi:10.1371/journal.pbio.0050057
43. KuriharaYMatsuiAKawashimaMKaminumaEIshidaJ
2008
Identification of the candidate genes regulated by RNA-directed
DNA methylation in Arabidopsis.
Biochem Biophys Res Commun
376
553
557
44. FuchsJDemidovDHoubenASchubertI
2006
Chromosomal histone modification patterns—from conservation
to diversity.
Trends Plant Sci
11
199
208
45. HuettelBKannoTDaxingerLAufsatzWMatzkeAJ
2006
Endogenous targets of RNA-directed DNA methylation and Pol IV in
Arabidopsis.
EMBO J
25
2828
2836
46. ZhangYReinbergD
2001
Transcription regulation by histone methylation: interplay
between different covalent modifications of the core histone
tails.
Genes Dev
15
2343
2360
47. SukaNSukaYCarmenAAWuJGrunsteinM
2001
Highly specific antibodies determine histone acetylation site
usage in yeast heterochromatin and euchromatin.
Mol Cell
8
473
479
48. MathieuOReindersJCaikovskiMSmathajittCPaszkowskiJ
2007
Transgenerational stability of the Arabidopsis epigenome is
coordinated by CG methylation.
Cell
130
851
862
49. MirouzeMReindersJBucherENishimuraTSchneebergerK
2009
Selective epigenetic control of retrotransposition in
Arabidopsis.
Nature
461
427
430
50. TeixeiraFKHerediaFSarazinARoudierFBoccaraM
2009
A role for RNAi in the selective correction of DNA methylation
defects.
Science
323
1600
1604
51. SlotkinRKVaughnMBorgesFTanurdzićMBeckerJD
2009
Epigenetic reprogramming and small RNA silencing of transposable
elements in pollen.
Cell
136
461
472
52. RobertsonKDAit-Si-AliSYokochiTWadePAJonesPL
2000
DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses
transcription from E2F-responsive promoters.
Nat Genet
25
338
342
53. RountreeMRBachmanKEBaylinSB
2000
DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a
complex at replication foci.
Nat Genet
25
269
277
54. BaiSGhoshalKDattaJMajumderSYoonSO
2005
DNA methyltransferase 3b regulates nerve growth factor-induced
differentiation of PC12 cells by recruiting histone deacetylase
2.
Mol Cell Biol
25
751
766
55. KatoMMiuraABenderJJacobsenSEKakutaniT
2003
Role of CG and non-CG methylation in immobilization of
transposons in Arabidopsis.
Curr Biol
13
421
426
56. ChanSWHendersonIRZhangXShahGChienJS
2006
RNAi, DRD1, and histone methylation actively target
developmentally important non-CG DNA methylation in
Arabidopsis.
PLoS Genet
2
e83
doi:10.1371/journal.pgen.0020083
57. JohnsonLMBostickMZhangXKraftEHendersonI
2007
The SRA methyl-cytosine-binding domain links DNA and histone
methylation.
Curr Biol
17
379
384
58. MathieuOProbstAVPaszkowskiJ
2005
Distinct regulation of histone H3 methylation at lysines 27 and 9
by CpG methylation in Arabidopsis.
EMBO J
24
2783
2791
59. OnoderaYHaagJReamTNunesPCPontesO
2005
Plant nuclear RNA polymerase IV mediates siRNA and
DNA-methylation-dependent heterochromatin formation.
Cell
120
613
622
60. MatsuiAIshidaJMorosawaTMochizukiYKaminumaE
2008
Arabidopsis transcriptome analysis under
drought, cold, high-salinity and ABA treatment conditions using a tiling
array.
Plant Cell Physiol
49
1135
1149
61. KimJMToTKIshidaJMorosawaTKawashimaM
2008
Alterations of lysine modifications on the histone H3 N-tail
under drought stress conditions in Arabidopsis
thaliana.
Plant Cell Physiol
49
1580
1588
62. KimuraHHayashi-TakanakaYGotoYTakizawaNNozakiN
2008
The organization of histone H3 modifications as revealed by a
panel of specific monoclonal antibodies.
Cell Struct Funct
33
61
73
63. SchmittgenTDLivakKJ
2008
Analyzing real-time PCR data by the comparative C(T)
method.
Nat Protoc
3
1101
1108
Štítky
Genetika Reprodukční medicína
Článek Incorporating Biological Pathways via a Markov Random Field Model in Genome-Wide Association StudiesČlánek Survival Motor Neuron Protein Regulates Stem Cell Division, Proliferation, and Differentiation inČlánek Epigenetic Regulation of Cell Type–Specific Expression Patterns in the Human Mammary Epithelium
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 4
-
Všechny články tohoto čísla
- Is Co-Expressed with Closely Adjacent Uncharacterised Genes Spanning a Breast Cancer Susceptibility Locus at 6q25.1
- The Liberation of Embryonic Stem Cells
- Genome-Wide Meta-Analysis Identifies Regions on 7p21 () and 15q24 () As Determinants of Habitual Caffeine Consumption
- A Sustained Dietary Change Increases Epigenetic Variation in Isogenic Mice
- The Exocyst Protein Sec10 Interacts with Polycystin-2 and Knockdown Causes PKD-Phenotypes
- Incorporating Biological Pathways via a Markov Random Field Model in Genome-Wide Association Studies
- Survival Motor Neuron Protein Regulates Stem Cell Division, Proliferation, and Differentiation in
- Identification and Functional Validation of the Novel Antimalarial Resistance Locus in
- Does Positive Selection Drive Transcription Factor Binding Site Turnover? A Test with Drosophila Cis-Regulatory Modules
- Protein Phosphatase 2A Controls Ethylene Biosynthesis by Differentially Regulating the Turnover of ACC Synthase Isoforms
- Ribosomal DNA Deletions Modulate Genome-Wide Gene Expression: “–Sensitive” Genes and Natural Variation
- Reciprocal Sign Epistasis between Frequently Experimentally Evolved Adaptive Mutations Causes a Rugged Fitness Landscape
- Variable Pathogenicity Determines Individual Lifespan in
- Evolution of Vertebrate Transient Receptor Potential Vanilloid 3 Channels: Opposite Temperature Sensitivity between Mammals and Western Clawed Frogs
- Towards Establishment of a Rice Stress Response Interactome
- Mouse Genome-Wide Association and Systems Genetics Identify As a Regulator of Bone Mineral Density and Osteoclastogenesis
- Quantitative Fitness Analysis Shows That NMD Proteins and Many Other Protein Complexes Suppress or Enhance Distinct Telomere Cap Defects
- Highly Precise and Developmentally Programmed Genome Assembly in Requires Ligase IV–Dependent End Joining
- PDP-1 Links the TGF-β and IIS Pathways to Regulate Longevity, Development, and Metabolism
- Genome-Wide Association Study Using Extreme Truncate Selection Identifies Novel Genes Affecting Bone Mineral Density and Fracture Risk
- Eight Common Genetic Variants Associated with Serum DHEAS Levels Suggest a Key Role in Ageing Mechanisms
- 14-3-3 Proteins Regulate Exonuclease 1–Dependent Processing of Stalled Replication Forks
- HDA6 Regulates Locus-Directed Heterochromatin Silencing in Cooperation with MET1
- Epigenetic Regulation of Cell Type–Specific Expression Patterns in the Human Mammary Epithelium
- Enhanced Statistical Tests for GWAS in Admixed Populations: Assessment using African Americans from CARe and a Breast Cancer Consortium
- Beyond Missing Heritability: Prediction of Complex Traits
- An Evolutionary Genomic Approach to Identify Genes Involved in Human Birth Timing
- Long-Lost Relative Claims Orphan Gene: in a Wasp
- PTG Depletion Removes Lafora Bodies and Rescues the Fatal Epilepsy of Lafora Disease
- Chromatin Organization in Sperm May Be the Major Functional Consequence of Base Composition Variation in the Human Genome
- GWAS of Follicular Lymphoma Reveals Allelic Heterogeneity at 6p21.32 and Suggests Shared Genetic Susceptibility with Diffuse Large B-cell Lymphoma
- Loss-of-Function Mutations in Cause Metachondromatosis, but Not Ollier Disease or Maffucci Syndrome
- DNA Damage, Somatic Aneuploidy, and Malignant Sarcoma Susceptibility in Muscular Dystrophies
- The Phylogenetic Origin of Coincided with the Origin of Maternally Provisioned Germ Plasm and Pole Cells at the Base of the Holometabola
- Genome Analysis Reveals Interplay between 5′UTR Introns and Nuclear mRNA Export for Secretory and Mitochondrial Genes
- Genome-Wide Association Analysis of Soluble ICAM-1 Concentration Reveals Novel Associations at the , , , and Loci
- The Complete Spectrum of Yeast Chromosome Instability Genes Identifies Candidate CIN Cancer Genes and Functional Roles for ASTRA Complex Components
- Dynamic Regulation of H3K27 Trimethylation during Differentiation
- Phosphorylation-Dependent Differential Regulation of Plant Growth, Cell Death, and Innate Immunity by the Regulatory Receptor-Like Kinase BAK1
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PTG Depletion Removes Lafora Bodies and Rescues the Fatal Epilepsy of Lafora Disease
- Evolution of Vertebrate Transient Receptor Potential Vanilloid 3 Channels: Opposite Temperature Sensitivity between Mammals and Western Clawed Frogs
- Survival Motor Neuron Protein Regulates Stem Cell Division, Proliferation, and Differentiation in
- An Evolutionary Genomic Approach to Identify Genes Involved in Human Birth Timing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání