-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaThe Histone Demethylase Jhdm1a Regulates Hepatic Gluconeogenesis
Hepatic gluconeogenesis is required for maintaining blood glucose homeostasis; yet, in diabetes mellitus, this process is unrestrained and is a major contributor to fasting hyperglycemia. To date, the impacts of chromatin modifying enzymes and chromatin landscape on gluconeogenesis are poorly understood. Through catalyzing the removal of methyl groups from specific lysine residues in the histone tail, histone demethylases modulate chromatin structure and, hence, gene expression. Here we perform an RNA interference screen against the known histone demethylases and identify a histone H3 lysine 36 (H3K36) demethylase, Jhdm1a, as a key negative regulator of gluconeogenic gene expression. In vivo, silencing of Jhdm1a promotes liver glucose synthesis, while its exogenous expression reduces blood glucose level. Importantly, the regulation of gluconeogenesis by Jhdm1a requires its demethylation activity. Mechanistically, we find that Jhdm1a regulates the expression of a major gluconeogenic regulator, C/EBPα. This is achieved, at least in part, by its USF1-dependent association with the C/EBPα promoter and its subsequent demethylation of dimethylated H3K36 on the C/EBPα locus. Our work provides compelling evidence that links histone demethylation to transcriptional regulation of gluconeogenesis and has important implications for the treatment of diabetes.
Published in the journal: . PLoS Genet 8(6): e32767. doi:10.1371/journal.pgen.1002761
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002761Summary
Hepatic gluconeogenesis is required for maintaining blood glucose homeostasis; yet, in diabetes mellitus, this process is unrestrained and is a major contributor to fasting hyperglycemia. To date, the impacts of chromatin modifying enzymes and chromatin landscape on gluconeogenesis are poorly understood. Through catalyzing the removal of methyl groups from specific lysine residues in the histone tail, histone demethylases modulate chromatin structure and, hence, gene expression. Here we perform an RNA interference screen against the known histone demethylases and identify a histone H3 lysine 36 (H3K36) demethylase, Jhdm1a, as a key negative regulator of gluconeogenic gene expression. In vivo, silencing of Jhdm1a promotes liver glucose synthesis, while its exogenous expression reduces blood glucose level. Importantly, the regulation of gluconeogenesis by Jhdm1a requires its demethylation activity. Mechanistically, we find that Jhdm1a regulates the expression of a major gluconeogenic regulator, C/EBPα. This is achieved, at least in part, by its USF1-dependent association with the C/EBPα promoter and its subsequent demethylation of dimethylated H3K36 on the C/EBPα locus. Our work provides compelling evidence that links histone demethylation to transcriptional regulation of gluconeogenesis and has important implications for the treatment of diabetes.
Introduction
Hepatic glucose production is critical for the maintenance of normal blood levels to meet whole-body fuel requirements. In the early phase of postabsorptive state, circulating glucose is supplied from breakdown of liver glycogen stores. When fasting progresses, gluconeogenesis, which utilizes non-carbohydrate precursors to de novo synthesize glucose, becomes the major form of hepatic glucose production [1], [2]. In both type 1 and type 2 diabetes, gluconeogenesis is exaggerated and contributes to hyperglycemia [3]–[5].
The rate of gluconeogenesis is largely determined by three rate-limiting enzymes, Phosphoenolpyruvate carboxykinase (PEPCK), fructose-1,6-bisphosphatase (FBP-1) and glucose 6-phosphatase (G6Pase). The levels of these gluconeogenic enzymes are controlled by hormonal signals, notably glucagon and glucocorticoids, and the opposing hormone insulin, at the transcription level. Key DNA elements responsible for the hormonal regulation have been well characterized on the promoters of PEPCK and G6Pase gene [6]–[9]. These elements serve as platforms for setting up a complex transcriptional machinery that includes transcription factors (e.g., CREB, FOXO1, FOXA2, C/EBPs, HNF4α, GR, Nur77) and co-factors (e.g., PGC-1α, CRTC2, SIRT1, p300/CBP, SRC-1), thereby driving gluconeogenic gene expression [10], [11]. Despite these tremendous progresses, the regulatory mechanisms upstream of this transcriptional network are incompletely understood. Furthermore, it is unclear how the chromatin landscape affects gluconeogenesis, what chromatin modifying enzymes (in addition to p300/CBP) are involved, and how these enzymes coordinate with the aforementioned transcriptional regulators.
One determinant for chromatin structure and functional state is histone methylation that occurs on specific lysine residues in histones [12], [13]. Five lysine residues within the N-terminal tail of histone H3 (K4, K9, K27, and K36) and H4 (K20) have been shown to be the sites for methylation. These lysine residues can be mono-, di-, or trimethylated. Depending on the specific lysine residues and the degree of methylation, histone methylation can have distinct effects on gene expression. In general, histone H3K4 and K36 di-and trimethylation, and H3K27 monomethylation are associated with actively transcribed genes, whereas H3K9 and K27 di - and trimethylation are considered repressive markers for gene expression. The distribution pattern of histone methylation on gene loci can also be quite different. For example, H3K4 and K9 methylation are enriched in the promoter regions, whereas K36 di - and trimethylation are mainly located in the coding regions and their levels peak toward the 3′end of the gene [14]–[16]. By altering chromatin structure, histone methylation fine-tunes transcriptional outputs.
Histone methylation is reversible and its dynamic nature is controlled by a balance between histone methyltransferases and histone demethylases. A number of histone demethylases have been identified in recent years and they are classified into two groups [17]–[20]. The first group contains two genes, LSD1 and LSD2, in human genome. These enzymes catalyze demethylation via an FAD-dependent oxidative reaction that requires protonated nitrogen in the substrate [19]. The second group are genes that contain a JmjC domain. Nineteen members of the JmjC domain-containing proteins in the human genome have been shown to be demethylases. The JmjC domain is the catalytic domain that possesses demethylation activity. These enzymes use Fe(II) and the intermediate metabolite α-ketoglutarate as co-factors to catalyze a hydroxylation-based demethylation [20]. Because of their enzymatic requirement for either FAD or α-ketoglutarate, it has been postulated that histone demethylases might be important for energy homeostasis by linking metabolic signals to chromatin status and transcriptional regulation [21]. Here, through both in vitro and in vivo studies, we reveal an important regulatory function of histone demethylase Jhdm1a in gluconeogenesis that is mediated by its active demethylation on the C/EBPα locus.
Results
Identification of the histone demethylase Jhdm1a as a negative regulator of gluconeogenic gene expression
To assess whether JmjC domain-containing histone demethylase(s) is involved in the regulation of gluconeogenesis, we treated human hepatoma HepG2 cells with N-oxalylglycine (NOG) or its derivative, dimethyloxalylglycine (DMOG), and examined the expression of the gluconeogenic enzymes. NOG and DMOG are analogues of α-ketoglutarate and are general enzymatic inhibitors of the JmjC domain-containing histone demethylases [22], [23]. Treatment with either compound led to an increase of PEPCK and G6Pase expression (Figure 1A), indicating a potential requirement of histone demethylation activity in the regulation of gluconeogenesis.
Fig. 1. Knockdown of Jhdm1a specifically upregulates PEPCK and G6Pase expression in cultured hepatic cells. 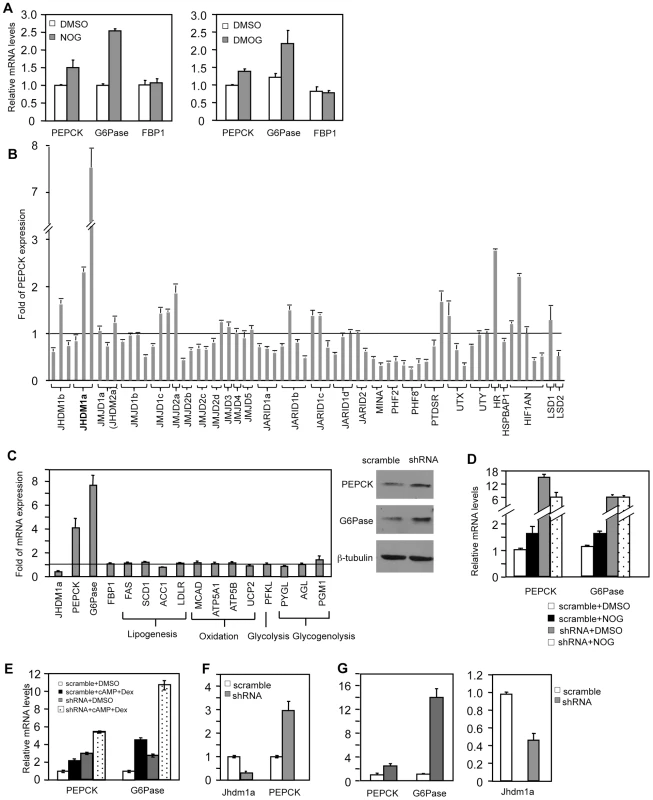
(A) HepG2 cells were treated with DMSO, NOG (1 mM), or DMOG (0.1 mM) for 12 hr. (B) shRNA-mediated screen. Each bar represents a single shRNA construct. Data were presented as fold relative to the scramble control. (C) Jhdm1a was knocked down in HepG2 cells with shRNA lentiviruses. Left, gene expression data were presented as fold relative to the scramble control from three experiments. Full names of individual genes are listed in Table S1. Right, levels of PEPCK and G6Pase protein were determined. (D) Jhdm1a knockdown or scramble HepG2 cells were treated with NOG (1 mM) for 12 hr. (E) Jhdm1a knockdown or scramble HepG2 cells were treated with a combination of dibutyryl cyclic-AMP (cAMP, 0.5 mM) and dexamethasone (Dex, 1 µM) in DMEM medium for 6 hr. Data were from two experiments. (F) Lentiviral knockdown of Jhdm1a in mouse hepatoma HepA1-6 cells. Data were from two experiments. (G) Adenoviral knockdown of Jhdm1a in mouse primary hepatocytes. Experiments were repeated three times with similar results. We next decided to use shRNA knockdown to identify the involved histone demethylase(s). As both the hormonal and molecular pathways that regulate PEPCK and G6Pase transcription are retained in HepG2 cells, we performed our screening experiments in these cells. We obtained a collection of human lentiviral shRNA constructs against the known JmjC domain-containing demethylases and a few JmjC domain-containing proteins where an enzymatic function has not been ascribed. We also included shRNA constructs against the FAD-dependent histone demethylases (LSD1 and LSD2). We stably expressed individual knockdown constructs in HepG2 cells and screened by quantitative RT-PCR for an increase of PEPCK expression compared with scramble controls. We found that knockdown of the JmjC-domain-containing protein Jhdm1a had the strongest effect (Figure 1B). Jhdm1a is a histone demethylase that specifically demethylates dimethylated H3K36 [20]. Knockdown of this demethylase also robustly promoted G6Pase expression, but not FBP-1 expression (Figure 1C). Not surprisingly, this led to an increase of PEPCK and G6Pase protein levels (Figure 1C). It is likely that NOG-induced PEPCK and G6Pase expression is mediated through inhibition of Jhdm1a, as the induction was lost in Jhdm1a knockdown cells (Figure 1D). As expected, treatment of HepG2 cells with dibutyryl cyclic-AMP and dexamethasone stimulated PEPCK and G6Pase expression; knockdown of Jhdm1a further led to an additive/synergistic increase, indicating a possibility that the effect of Jhdm1a knockdown is independent of the pathway activated by the hormones (Figure 1E). Similar results were obtained in HepG2 cells with an independent Jhdm1a silencing construct (Figure S1). In addition, we generated a lentiviral knockdown construct that targeted mouse Jhdm1a and expressed it in mouse hepatoma HepA1-6 cells. These cells express low level of PEPCK and undetectable level of G6Pase. Silencing of Jhdm1a in these cells elevated PEPCK expression (Figure 1F), while expression of key gluconeogenic transcriptional regulator Foxo1 and PGC-1α was not increased (Figure S2). We next studied gluconeogenic gene expression in a more physiological setting. We knocked down Jhdm1a in primary mouse hepatocytes using adenovirus and found that PEPCK and G6Pase expression was increased as well (Figure 1G). These results collectively demonstrated a negative role of Jhdm1a in gluconeogenic gene expression. Finally, we determined whether Jhdm1a regulates other metabolic pathways in HepG2 cells. We found that knockdown of Jhdm1a did not affect expression of any examined genes involved in lipogenesis, fatty acid oxidation, glycolysis, or glycogenolysis (Figure 1C), suggesting a quite specific metabolic function of Jhdm1a.
Ectopic expression of Jhdm1a suppresses gluconeogenic gene expression in a demethylation activity-dependent manner
Given that knockdown of Jhdm1a elevates gluconeogenic gene expression, we examined whether an opposite effect could be observed in cells expressing Jhdm1a. We stably expressed Jhdm1a via lentivirus in liver cells and found that this expression decreased both basal and hormonal-stimulated levels of PEPCK and G6Pase mRNA (Figure 2A and Figure S3). Interestingly, and in agreement with the knockdown data (Figure 1B), ectopic expression of demethylase Jhdm1b, which is closely related to Jhdm1a, did not inhibit gluconeogenic gene expression (Figure 2B and Figure S4). To determine the domains in Jhdm1a that are required for its suppressive function, we generated a series of Jhdm1a mutants. We first confirmed that these mutants were capable of producing stable proteins at a similar level, as judged by plasmid transfection in Hela cells (Figure 2A). We then expressed the mutants in HepG2 cells through lentivirus with a similar, low infection efficiency. Deletion of the JmjC domain or the CXXC Zinc finger domain abolished the suppression on PEPCK and G6Pase expression, whereas mutant lacking either the PHD domain or the F-box and Leucine-rich repeats remained fully functional (Figure 2A and Figure S3). Note that these Jhdm1a mutants were expressed at similar mRNA levels as their wild-type counterpart. The JmjC domain harbors the histone demethylation activity. Consistent with the effect of the JmjC deletion mutant, a demethylation-dead point mutant (H212A) [20] of Jhdm1a was no longer able to suppress PEPCK and G6Pase expression (Figure 2A and Figure S3). We next determined the effect of Jhdm1a on glucose production in vitro. We found that ectopic expression of wild type Jhdm1a, but not the demethylation defective mutants, inhibited glucose production in rat hepatoma FAO cells (Figure 2C). Taken together, these results demonstrate that both the demethylation activity and the CXXC Zinc finger domain of Jhdm1a are required for its negative modulation of gluconeogenic gene expression.
Fig. 2. Suppression of gluconeogenic gene expression by Jhdm1a requires its demethylation activity. 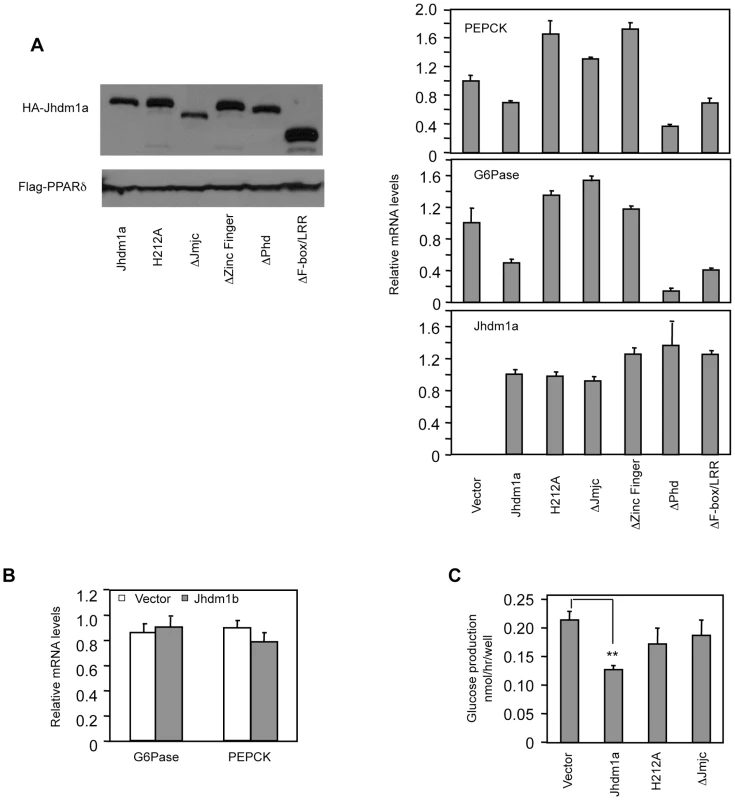
(A) (Left panel) Jhdm1a constructs were tagged with HA and transfected into Hela cells to ensure they express stable proteins. Co-transfected Flag-PPARδ serves as transfection and loading control. (Right, three panels) Jhdm1a constructs were lentivirally expressed in HepG2 cells. Data were shown from one representative of five experiments with similar results. (B) Lentiviral expression of Jhdm1b does not suppress gluconeogenic gene expression. (C) Jhdm1a constructs were lentivirally expressed in rat hepatic FAO cells and glucose production was measured. Data were from triplicates. **, P<0.01. Jhdm1a regulates gluconeogenesis in vivo
Based on our discovery of the regulatory role of Jhdm1a in vitro, we tested whether Jhdm1a regulates gluconeogenesis in live animals. We obtained five lentiviral Jhdm1a knockdown constructs from Open Biosystems and tested their knockdown efficiency by RT-QPCR in mouse cell culture. We transferred two best ones into an adenoviral vector, generated adenoviruses, and further confirmed that they were able to reduce ectopically expressed Jhdm1a protein level in vitro (Figure S5). The viruses were infused into the liver of wild-type C57BL/6J mice via tail vein injection and endogenous Jhdm1a expression was decreased, which led to a significantly increase in hepatic expression of PEPCK and G6Pase in both fed and fasting states, compared with the scramble control (Figure 3A, Figure S6 and S7). A corresponding enhanced PEPCK and G6Pase protein production was observed (Figure 3A). Blood insulin levels examined at fed state were not significantly different (Figure 3B). Although the Jhdm1a knockdown mice were still able to maintain normal glycemia, they displayed higher glucose production upon injection of the gluconeogenic substrate pyruvate (Figure 3C). We next ectopically expressed either wild-type Jhdm1a or the H212A point mutant in the liver of diabetic ob/ob mice. Expression of the wild-type Jhdm1a, but not the H212A point mutant, decreased the expression of PEPCK and G6Pase (Figure 3D). Accordingly, we observed a statistically significant reduction of blood glucose level in ob/ob mice expressing wild-type Jhdm1a (Figure 3E). Thus, Jhdm1a indeed has a physiological role in hepatic gluconeogenesis in vivo, and this role is mediated by its histone demethylation activity.
Fig. 3. Jhdm1a regulates gluconeogenesis in vivo. 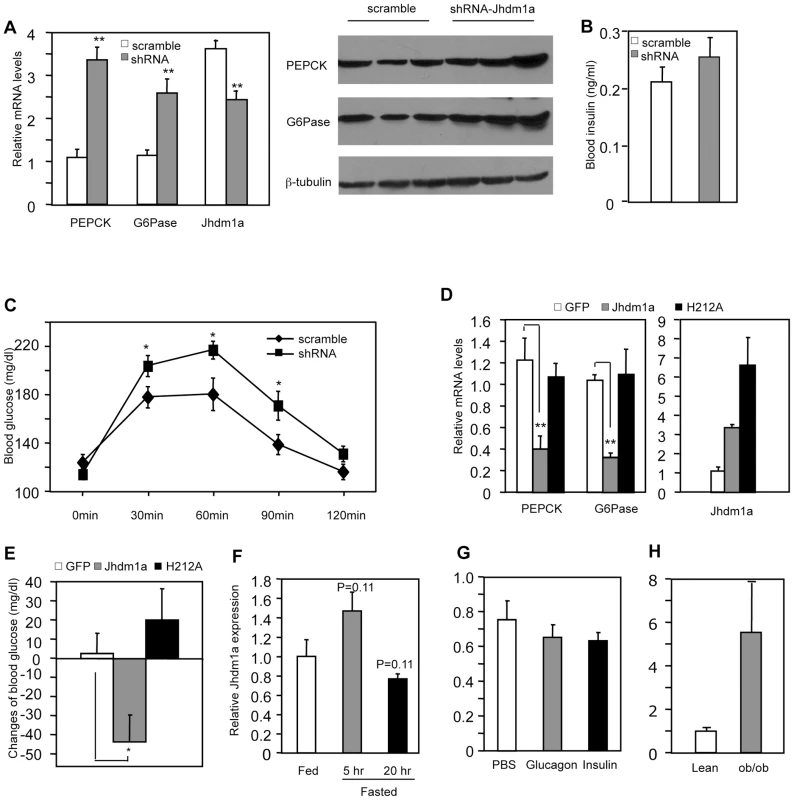
(A) Jhdm1a knockdown or scramble adenoviruses were transduced into the liver of wild-type male C57BL/6J mice (n = 5 per group). Mice fed ad libitum were sacrificed at Day 5 after viral infusion. (Left) mRNA levels of PEPCK, G6Pase and Jhdm1a in the liver were measured and normalized to U36b4. **, P<0.005. (Right) PEPCK and G6Pase protein. (B) Blood insulin levels at fed state were measured at Day 5. (C) Jhdm1a knockdown or scramble adenoviruses were transduced into the liver of wild-type male C57BL/6J mice (n = 10 per group). At Day 5, mice were i.p. injected with pyruvate (2 g/kg body weight) after a starvation for 16 hr and blood glucose levels were measured. *, P<0.05. (D and E) Adenoviruses expressing wild-type Jhdm1a, H212A point mutant, or GFP were transduced into the liver of male ob/ob mice (n = 5 per group). Gene expression was measured on Day 5 and blood glucose levels were measured on Day 3 after a 5-hr fasting. Changes of blood glucose level relative to Day 0 are presented. *, P<0.03; **, P<0.01. (F) Hepatic Jhdm1a mRNA levels in male C57BL/6J mice (n = 5 per group) fed ad libitum, or fasted for 5 hr or 20 hr. (G) Male C57BL/6J mice (n = 4) were i.p. injected with glucagon (300 µg/kg), insulin (0.75 U/Kg), or PBS. Hepatic Jhdm1a mRNA levels were examined 6 hr after injection. (H) Hepatic Jhdm1a mRNA levels in lean mice and diabetic ob/ob mice (n = 3 per group). Gluconeogenesis is activated during fasting and suppressed by a meal. Interestingly, the hepatic expression Jhdm1a was not changed during either a short-fasting (5 hr) or a long-fasting (20 hr) (Figure 3F). Furthermore, administration of either glucagon or insulin in vivo revealed no difference in Jhdm1a expression (Figure 3G). Likewise, treatment of HepG2 cells with dibutyryl cyclic-AMP and dexamethasone or insulin had no effect on Jhdm1a expression (Figure S8). Although we cannot rule out the possibility of post-transcriptional regulation of Jhdm1a by hormonal signaling, these data, together with the observed effects of Jhdm1a on PEPCK and G6Pase expression in both non-stimulatory and stimulatory conditions (Figure 1C and 1E, Figure 3A, and Figures S6 and S7), indicate that Jhdm1a acts as a negative regulatory mechanism to fine-tune baseline gluconeogenesis. In diabetic ob/ob mice, Jhdm1a expression was elevated (Figure 3H), possibly reflecting a feedback response.
Regulation of gluconeogenesis by Jhdm1a is mediated through C/EBPα expression
We explored how Jhdm1a regulates gluconeogenesis. We initially speculated that Jhdm1a might associate with the transcriptional regulator complex on the promoters of PEPCK and G6Pase and directly regulate their expression. To test this idea, we performed chromatin immunoprecipitation experiments in HepG2 cells ectopically expressing HA-tagged Jhdm1a. Unexpectedly, Jhdm1a did not associate with either PEPCK promoter or G6Pase promoter (Figure S9). The promoter regions we examined have been well characterized previously and are subjected to extensive regulation by an array of transcription regulators [6]–[9]. The lack of association of Jhdm1a with PEPCK and G6Pase promoters indicates to us that Jhdm1a might not directly regulate the expression of these two genes. We thus considered a possibility that Jhdm1a instead regulates the expression of any of the involved transcription factors or co-factors [10], [11]. We knocked down Jhdm1a in HepG2 cells and examined their expression. We found that the transcription factor C/EBPα was the only one whose expression level was significantly increased (Figure 4A). Similarly, knockdown of Jhdm1a promoted C/EBPα expression in primary mouse hepatocytes (Figure 4B). As a result of increased C/EBPα level, the association of C/EBPα with its binding sites within the PEPCK and G6Pase promoters was strongly enhanced in Jhdm1a knockdown HepG2 cells (Figure 4C). Members of C/EBPs were shown to activate the expression of PEPCK and G6Pase in vitro [24], [25]. We confirmed these previous results and also observed a remarkably similar target gene expression pattern between Jhdm1a silencing and C/EBPα ectopic expression (comparing Figure 1C and Figure 4D), supporting a functional connection between Jhdm1a and C/EBPα.
Fig. 4. Jhdm1a regulates the expression of C/EBPα, thereby indirectly modulating gluconeogenic gene expression. 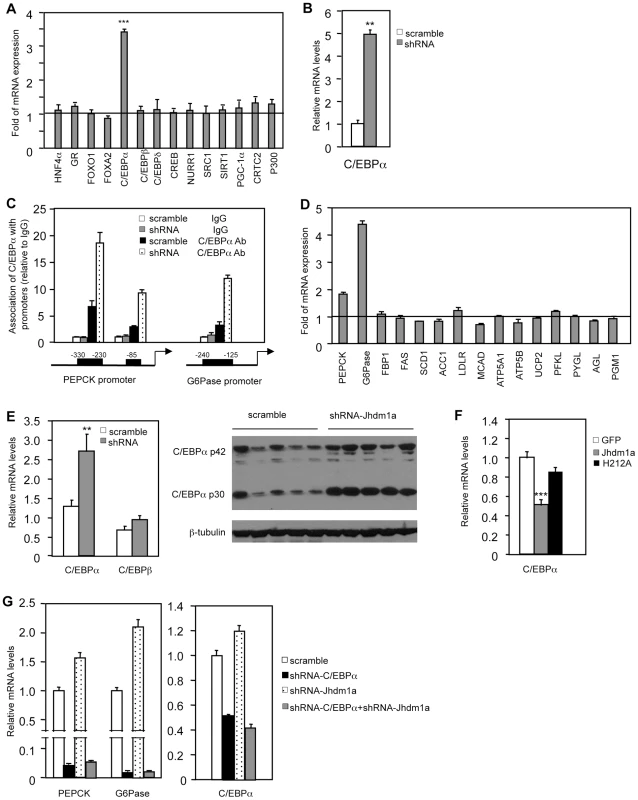
(A) Jhdm1a was knocked down in HepG2 cells with shRNA lentiviruses. Expression levels of known transcriptional regulators for gluconeogenesis were examined. Data are presented as fold relative to the scramble control from three experiments. ***, P<0.00005. (B) C/EBPα expression in Jhdm1a knockdown mouse primary hepatocytes. (C) Jhdm1a was knocked down in HepG2 cells with shRNA lentiviruses. Endogenous C/EBPα association with known binding sites on the PEPCK and G6Pase promoters was examined by ChIP assay. (D) Gene expression in HepG2 cells infected with lentiviruses expressing C/EBPα or vector. (E) Increased C/EBPα expression in the liver of wild-type C57BL/6J mice (n = 5 per group) with Jhdm1a knockdown. C/EBPα mRNA level and protein level were shown from independent groups of mice. **, P<0.02. (F) Decreased C/EBPα expression in the liver of ob/ob mice (n = 5 per group) ectopically expressing wild-type Jhdm1a, but not in the liver expressing H212A point mutant. ***, P<0.001. (G) HepG2 cells were infected with lentiviruses expressing C/EBPα shRNA and selected with puromycine. Cells were then infected with lentiviruses expressing Jhdm1a shRNA without selection. Data were shown from one representative of four experiments. Note, the low induction of PEPCK and G6Pase expression by Jhdm1a knockdown is due to the lack of selection pressure. Previous work by others has also demonstrated an essential in vivo role of C/EBPα in hepatic PEPCK and G6Pase expression [26]–[29]. Importantly, we found that in vivo knockdown of Jhdm1a in the mouse liver increased the level of C/EBPα (Figure 4E and Figure S10). Conversely, exogenous expression of Jhdm1a in the liver suppressed C/EBPα expression, whereas the H212A point mutant had no effect (Figure 4F). To further examine whether the action of Jhdm1a is C/EBPα-dependent, we knocked down both Jhdm1a and C/EBPα in hepatic cells. The increase of PEPCK and G6Pase expression caused by Jhdm1a knockdown was greatly diminished in the double knockdown cells (Figure 4G). The results together suggest that Jhdm1a regulates gluconeogenesis, at least in part, through its control of C/EBPα expression. As previously noted [11], C/EBPα expression remained unchanged during both short and long fasting (data not shown), in agreement with our observation that Jhdm1a expression was not affected by these conditions.
USF1 mediates the recruitment of Jhdm1a to the C/EBPα promoter
To identify the molecular mechanism by which Jhdm1a regulates C/EBPα expression, we first examined whether Jhdm1a associates with the C/EBPα locus. The C/EBPα locus contains a single exon. We expressed HA-tagged Jhdm1a in hepatic cells and performed chromatin immunoprecipitation experiments using antibody against the HA tag. We found that Jhdm1a was associated with the C/EBPα promoter region but not with the intragenic region (Figure 5A). Interestingly, this promoter region contains four separate transcription factor USF1 binding sites that have been implicated in C/EBPα expression [30], [31] and Jhdm1a was present on three of them. An interaction between Jhdm1a and USF1 was readily detected in cells expressing both of them (Figure 5B). Moreover, reduction of USF1 level by shRNA-mediated silencing diminished the association of exogenous Jhdm1a with these sites (Figure 5C). Despite the high background of the Jhdm1a antibody, we were also able to show that endogenous Jhdm1a associated with the USF1 binding sites, since knockdown of Jhdm1a decreased its association with these sites (Figure S11). Functionally, knockdown of USF1 led to an increase of C/EBPα expression and accordingly, an increase of PEPCK expression (Figure 5D). These data suggest a model in which USF1 recruits Jhdm1a to the C/EBPα promoter to negatively regulate its expression.
Fig. 5. Suppression of C/EBPα expression by Jhdm1a is mediated by USF1. 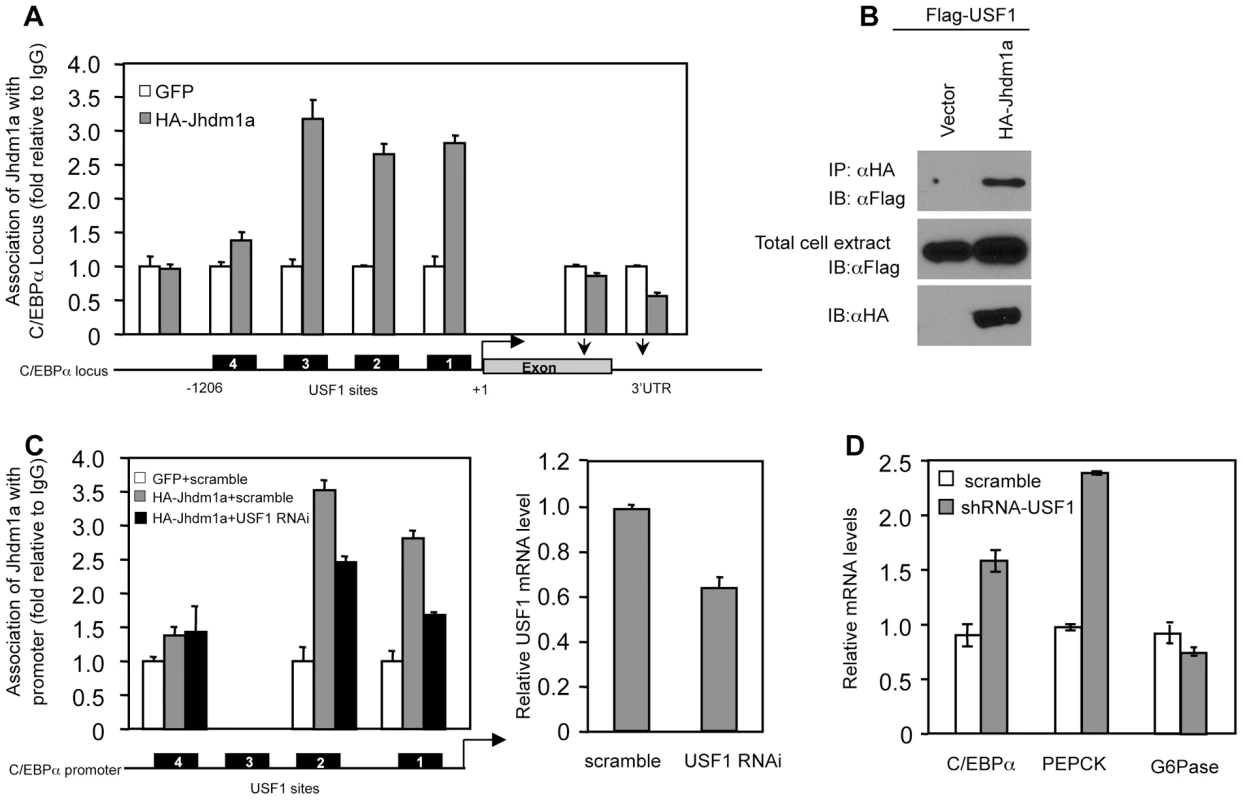
(A) Jhdm1a associates with putative USF1-binding sites on the C/EBPα promoter region. Adenoviral HA-Jhdm1a were expressed in HepG2 cells and ChIP assays were performed with HA antibody. Data were shown from one representative of three experiments with similar results. (B) Jhdm1a interacts with USF1. Hela cells were co-transfected with indicated plasmids. Cell extracts were incubated with HA beads and immunoprecipitates were probed with Flag antibody. (C) HA-Jhdm1a (adenoviral) along with shRNA (lentiviral) against USF1 was co-expressed in HepG2 cells. ChIP assays were performed with HA antibody. (D) Gene expression in HepG2 cells expressing lentiviral USF1 shRNA. Data were shown from one representative of three experiments. Jhdm1a actively demethylates dimethylated H3K36 on the C/EBPα locus
Given the association of Jhdm1a with the C/EBPα promoter, we examined whether Jhdm1a modulates the H3K36 methylation pattern on the C/EBPα locus. Knockdown of Jhdm1a increased H3K36 dimethylation in the 3′ exon region and 3′ UTR that is close to the exon, but had little effect on H3K36 dimethylation on the promoter, 5′ exon region, and 3′UTR that is located far away from the exon (Figure 6A). This pattern of modulation is in concord with the previously shown genome-wide distribution of H3K36 dimethylation where it is mostly found in the intragenic region and usually peaks toward 3′ exon [15]. The demethylation by Jhdm1a is gene-specific, as knockdown of Jhdm1a did not increase H3K36 dimethylation at the C/EBPβ locus (Figure 6B). Moreover, knockdown of Jhdm1a did not affect the H3K36 trimethylation pattern at the C/EBPα locus (Figure 6C), consistent with the enzymatic property of Jhdm1a to specifically demethylate dimethylated H3K36 [20]. Next, we examined the effect of ectopically expressed Jhdm1a on H3K36 dimethylation at the C/EBPα locus. We found that expression of wild type Jhdm1a, but not of the H212A mutant, led to a significant decrease of K36 dimethylation (Figure 6D). These results suggest that Jhdm1a demethylates dimethylated H3K36 at the C/EBPα locus, hence directly regulating its expression.
Fig. 6. Jhdm1a specifically demethylates dimethylated H3K36 on the C/EBPα locus. 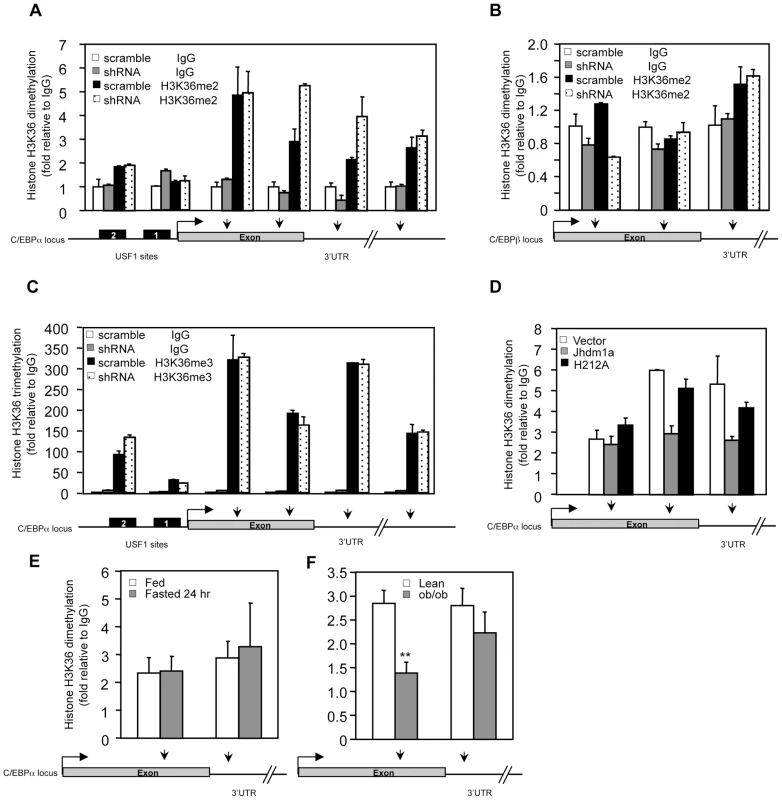
(A) Knockdown of Jhdm1a increases H3K36 dimethylation (H3K36me2) on the C/EBPα locus. Data were shown from one representative of three independent experiments with similar results. (B) Knockdown of Jhdm1a does not affect H3K36me2 on the C/EBPβ locus. (C) Knockdown of Jhdm1a does not increase H3K36 trimethylation (H3K36me3) on the C/EBPα locus. (D) Wild-type Jhdm1a, but not the H212A point mutant, decreases H3K36me2 on the C/EBPα locus. (E) H3K36me2 on the C/EBPα locus is not modulated by feeding/fasting conditions. n = 4 per group. (F) Decreased H3K36me2 on the C/EBPα locus in ob/ob mice. n = 4 per group. **, P<0.01. We determined whether the H3K36 dimethylation status at the C/EBPα locus is modulated by hormonal signaling or metabolic states. In agreement with Jhdm1a expression (Figure 3F and 3G, and Figure S8), we found that levels of H3K36 dimethylation remained unchanged in HepG2 cells treated with hormones (Figure S12) or in livers of fasted mice (Figure 6E), supporting the idea that Jhdm1a and H3K36 dimethylation at the C/EBPα locus are primarily involved in basal control of gluconeogenesis. Interestingly, H3K36 dimethylation was significantly decreased at the exon region of C/EBPα locus in diabetic ob/ob mice (Figure 6F), likely due to increased Jhdm1a expression (Figure 3H). These data suggest a possible physiological, compensatory attempt to suppress hyperglycemia in ob/ob mice.
Discussion
In recent years, a number of histone demethylases have been identified [17]–[20]. While these exciting discoveries dramatically reversed our previous view that histone methylation was a stable, non-erasable marker, our knowledge regarding the functions of these demethylases in biological processes and diseases is very limited. Here, through an shRNA screen against the known histone demethylases, we identify Jhdm1a negatively regulates gluconeogenic gene PEPCK and G6Pase expression both in vitro and in vivo. Phenotypically, silencing of Jhdm1a elevates glucose production, whereas its ectopic expression lowers blood glucose levels in diabetes. Interestingly, our studies suggest that Jhdm1a does not appear to control PEPCK and G6Pase expression directly. Rather, Jhdm1a exerts its function through C/EBPα. The role of C/EBPα in gluconeogenesis has been well established [25]–[29]. We found that Jhdm1a negatively modulates the expression of C/EBPα through active demethylation on the C/EBPα locus. Therefore, our work potentially uncovers a novel molecular mechanism in gluconeogenesis, where histone demethylation regulates a key gluconeogenic transcription factor. However, it is important to note that our in vivo studies were performed using adenoviral infusion to acutely manipulate hepatic Jhdm1a level, therefore, chronic and more physiological and pathophysiological roles of Jhdm1a in gluconeogenesis remain to be addressed in detail with liver-specific Jhdm1a knockout and transgenic models. In addition, as genetic variations at the Jhdm1a locus are present in human population, it will be interesting to analyze whether these variations are associated with type 2 diabetes.
It was hypothesized that histone demethylases might be important for metabolic homeostasis [21]. This is supported by the obese phenotype of mice deficient for H3K9 histone demethylase, Jhdm2a [32], [33]. Our demonstration of Jhdm1a functioning in gluconeogenesis provides another example. It is anticipated that future studies will reveal additional histone demethylases as important regulators of energy metabolism. Histone demethylases are considered as global modifiers of chromatin structure, however, it is clear that a particular demethylase only regulates a small subset of genes and therefore, a specific metabolic pathway. This specificity is likely to be determined by the target tissue, the repertoire of transcriptional regulators in that tissue, and whether this particular demethylation on individual gene locus is sufficient to translate into a gene expression readout.
Histone H3K36 di - and trimethylation have been shown to be associated with actively transcribed genes and their levels peak near the 3′ end of the gene [14]–[16], [34]. In yeast, K36 di - and trimethylation have been implicated in transcriptional elongation by preventing cryptic, intragenic transcription [35]–[37]. In higher eukaryotes, the exact function of K36 methylation is poorly understood. We show here that Jhdm1a demethylates dimethylated H3K36 on the C/EBPα locus and negatively regulates its expression. Although we cannot rule out the possibility that changes of dimethylated H3K36 level are secondary due to C/EBPα expression, the requirement for the demethylase activity of Jhdm1a and the unaffected H3K36 trimethylation on the C/EBPα locus strongly argue that this is unlikely. Jhdm1b is another demethylase that targets dimethylated H3K36. Jhdm1b-mediated demethylation was recently shown to negatively regulate the expression of the p15Ink4b tumor suppressor [38]. These studies suggest a positive role of H3K36 dimethylation for gene expression.
Our data suggest that Jhdm1a is recruited by USF1 to the USF1-binding sites within the C/EBPα promoter. A recent study shows that Jhdm1a, through its CXXC Zinc finger domain, associates with unmethylated CpG islands on gene promoters [39]. Indeed, the C/EBPα promoter is considerably CpG-rich, and we find that the CXXC Zinc finger domain is required for the suppressive function of Jhdm1a. Therefore, it is possible that the CpG-rich sequences and USF1 cooperatively mediate the recruitment of Jhdm1a. One interesting observation in our study is that Jhdm1a demethylates C/EBPα intragenic region that lacks detectable association. It is possible that the initial recruitment by USF1 to the promoter is a relatively stable state, but following recruitment, Jhdm1a moves along the gene body to demethylate dimethylated H3K36. Thus the association of Jhdm1a with the gene body might be transient and difficult to capture. There are precedents of similar observations. For example, ChIP-seq studies reveal that, for actively transcribed genes, Pol II is predominantly detected at transcription start sites, not transcribed regions [16]. PHF8, a H4K20/H3K9 demethylase, was found to demethylate regions that it does not associate with [40]. Clearly, how epigenetic enzymes are recruited and are able to modify chromatin structure in a widespread fashion is a fascinating question to be fully understood.
While Jhdm1a-catalyzed histone demethylation regulates gluconeogenesis through an indirect mechanism by targeting C/EBPα, a previous report has postulated that dimethylation of histone H3 arginine 17 has a direct impact on gluconeogenic gene expression, as the level of this modification on the PEPCK promoter increases with dexamethasone treatment and decreases upon subsequent addition of insulin [41]. However, the molecular events responsible for and the functional outcome of this change were unknown. Nevertheless, their studies, along with ours, indicate that histone methylation/demethylation could be more commonly employed than we appreciated to regulate gluconeogenesis at multiple layers. To our surprise, Jhdm1a expression, hence the H3K36 dimethylation status at the C/EBPα locus, are not influenced by fed and fasted states and hormonal signaling. Our data indicate that, under normal conditions, Jhdm1a-mediated demethylation primarily function in maintaining basal-state gluconeogenesis irrespective of nutritional and hormonal cues. In support of this model, we found that knockdown of Jhdm1a in mice elevates the expression of C/EBPα, PEPCK and G6Pase in both fed and fasted states. Mechanisms controlling hormonal-regulated gluconeogenesis have been extensively studies [11], less was understood for basal-state gluconeogenesis. Our work provides insights into this key process. Interestingly, in diabetic state, Jhdm1a expression is increased and H3K36 dimethylation at the C/EBPα locus is decreased, indicating a possible involvement of Jhdm1a in counteracting hyperglycemia. Thus, under pathophysiological conditions such as obesity and insulin resistance, the expression and/or activity of Jhdm1a can be modulated by currently unknown mechanisms. In addition, we find that in fetal liver, Jhdm1a is highly expressed and C/EBPα level is very low; in neonatal stage, hepatic Jhdm1a level decreases and C/EBPα level increases (our unpublished data). As gluconeogenesis occurs in neonatal stage but not in embryonic stage, whether Jhdm1a is involved in this metabolic transition during development remains to be determined. In summary, our results illustrate how the dynamics of H3K36 dimethylation regulates basal gluconeogenesis and indicate that increasing the demethylation activity of Jhdm1a could potentially offer therapeutic benefits to curb hyperglycemia.
Materials and Methods
Lentiviral knockdown
Lentiviral shRNA constructs (pGIPZ-based; Open Biosystems) against the known human demethylases were obtained through the RNAi Core Facility at University of Massachusetts Medical School. All other lentiviral shRNA constructs were obtained directly from Open Biosystems. All relevant constructs were verified and their targeting sequences are provided in Table S1. Lentiviruses were packaged as described [42]. After virus infection, cells were re-plated next day and selected with puromysin for three days. Cells were then trypsinized and plated at a similar confluency. Cells were cultured in the presence of puromycin for two more days and total RNA was isolated.
Lentiviral overexpression
Mouse wild-type and mutant Jhdm1a expression plasmids were generated by standard procedure and were fully sequenced. They were then transferred to pENTR-1A vector and recombined with pLenti-CMV/neo to generate lentiviral constructs essentially as described [43]. The titers of packaged lentiviruses were determined in liver cells. Cells were infected with similar number of viral particles, selected with G418, and cultured as above.
Gene expression
Total RNA was extracted with Trizol reagent. Gene expression was measured by quantitative RT-PCR and normalized to internal control genes (β-actin for cells, U36b4 or cyclophilin for liver tissue). Primer sequences are provided in Table S1.
Glucose production
Rat hepatic FAO cells expressing lentiviral Jhdm1a constructs were washed 3 times with PBS and then incubated in glucose free DMEM medium containing 2 mM sodium pyruvate and 20 mM sodium lactate for 6 hr. Glucose levels in the medium were measured with a Amplex red glucose assay kit (Invitrogen, #A22189).
Primary mouse hepatocytes
Cells were prepared and cultured as described [24]. Cells were infected with adenoviruses at a multiplicity of infection of 50. Two days after infection, cells were starved for 6 hr in DMEM supplemented with 0.2% BSA and 2 mM sodium pyruvate before RNA isolation.
Adenoviral infusion and animal studies
Adenoviral Jhdm1a expression and knockdown constructs and their respective control constructs were generated, and adenoviruses were produced and purified as described [42], [44]. Viral titers were determined in HEK293 cells by scoring GFP positive cells. Male wild-type C57BL/6J and ob/ob (on C57BL/6J background) mice were obtained from The Jackson Laboratory. Adenoviruses (4×109 and 9×109 viral particles for expression and knockdown, respectively) suspended in 0.2 ml PBS were injected through tail vein when animals were 10-week-old. Blood glucose levels were measured at indicated time and animals were sacrificed at Day 5. For pyruvate tolerance test, mice were fasted for 16 hr and sodium pyruvate dissolved in PBS was i.p. injected (2 g/kg body weight).
To determine the levels of liver PEPCK, G6Pase and C/EBPα protein, 50 mg liver sample were homogenized in 1 ml lysis buffer [100 mM NaCl, 50 mM Tris (pH 7.5), 0.5% Triton X-100, 5% (w/v) glycerol]. 26 µg protein extracts were separated by SDS-PAGE and probed with antibody against C/EBPα (Santa Cruz, sc-61), PEPCK (ABcam, ab28455) or G6Pase (Santa Cruz, sc-25840).
Co-immunoprecipitation
HA-Jhdm1a and Flag-USF1 plasmids were co-transfected into Hela cells. Cells were lysed in buffer [100 mM NaCl, 50 mM Tris (pH 7.5), 0.5% Triton X-100, 5% (w/v) glycerol]. Cell extracts were incubated with anti-HA beads (Santa Cruz, sc-7392AC) for overnight and the beads were washed 4 times with buffer [100 mM NaCl, 50 mM Tris (pH 7.5), 0.1% Triton X-100, 5% glycerol]. Immunoprecipitates were probed with an anti-Flag antibody (Sigma, F7425).
ChIP assays
Assays were performed as described [42] using antibodies against HA (Sigma, #H6908), C/EBPα (Santa Cruz, sc-61), dimethyl-H3K36 (Millipore, #07274), trimethyl-H3K36 (Abcam, #9050). Immunoprecipitate signal was normalized with input signal; both were measured by real-time QPCR. Primer sequences are provided in the Table S1.
For ChIP assays performed with liver samples, samples were generated as described with minor modifications [45]. Briefly, parts of liver from same locations were excised, cut into small pieces with a razor blade, cross-linked with 1% formaldehyde for 15 minutes at room temperature. The samples were then ground and filtered through a 40 µm cell strainer to produce a single liver cell suspension. Nuclear extracts were prepared, chromatin was sonicated using a ultrasonic processor, and immunoprecipitation was performed as described [42]. An equivalent of 40 mg of liver tissue was used for each immunoprecipitaiton. After normalized with inputs, ChIP signals were calculated as folds relative to background signal (IgG) generated from the same animal.
Statistical analysis
Student's t test (two-tailed) was used for statistical analysis. P<0.05 was considered significant. Data are presented as mean ± s.e.m.
Supporting Information
Zdroje
1. NordlieRCFosterJDLangeAJ 1999 Regulation of glucose production by the liver. Annu Rev Nutr 19 379 406
2. PilkisSJGrannerDK 1992 Molecular physiology of the regulation of hepatic gluconeogenesis and glycolysis. Annu Rev Physiol 54 885 909
3. MagnussonIRothmanDLKatzLDShulmanRGShulmanGI 1992 Increased rate of gluconeogenesis in type II diabetes mellitus. A 13C nuclear magnetic resonance study. J Clin Invest 90 1323 1327
4. WajngotAChandramouliVSchumannWCEkbergKJonesPK 2001 Quantitative contributions of gluconeogenesis to glucose production during fasting in type 2 diabetes mellitus. Metabolism 50 47 52
5. KunertOStinglHRosianEKrssakMBernroiderE 2003 Measurement of fractional whole-body gluconeogenesis in humans from blood samples using 2H nuclear magnetic resonance spectroscopy. Diabetes 52 2475 2482
6. ChakravartyKCassutoHReshefLHansonRW 2005 Factors that control the tissue-specific transcription of the gene for phosphoenolpyruvate carboxykinase-C. Crit Rev Biochem Mol Biol 40 129 154
7. RoeslerWJ 2000 What is a cAMP response unit? Mol Cell Endocrinol 162 1 7
8. Vander KooiBTOnumaHOeserJKSvitekCAAllenSR 2005 The glucose-6-phosphatase catalytic subunit gene promoter contains both positive and negative glucocorticoid response elements. Mol Endocrinol 19 3001 3022
9. LinBMorrisDWChouJY 1997 The role of HNF1alpha, HNF3gamma, and cyclic AMP in glucose-6-phosphatase gene activation. Biochemistry 36 14096 14106
10. AltarejosJYMontminyM 2011 CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol 12 141 151
11. LinHVAcciliD 2011 Hormonal regulation of hepatic glucose production in health and disease. Cell Metab 14 9 19
12. KouzaridesT 2007 Chromatin modifications and their function. Cell 128 693 705
13. MartinCZhangY 2005 The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6 838 849
14. EdmundsJWMahadevanLCClaytonAL 2008 Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J 27 406 420
15. BannisterAJSchneiderRMyersFAThorneAWCrane-RobinsonC 2005 Spatial distribution of di - and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 280 17732 17736
16. BarskiACuddapahSCuiKRohTYSchonesDE 2007 High-resolution profiling of histone methylations in the human genome. Cell 129 823 837
17. AggerKChristensenJCloosPAHelinK 2008 The emerging functions of histone demethylases. Curr Opin Genet Dev 18 159 168
18. MosammaparastNShiY 2010 Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem 79 155 179
19. ShiYLanFMatsonCMulliganPWhetstineJR 2004 Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119 941 953
20. TsukadaYFangJErdjument-BromageHWarrenMEBorchersCH 2006 Histone demethylation by a family of JmjC domain-containing proteins. Nature 439 811 816
21. TeperinoRSchoonjansKAuwerxJ 2010 Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab 12 321 327
22. AllegrucciCWuYZThurstonADenningCNPriddleH 2007 Restriction landmark genome scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum Mol Genet 16 1253 1268
23. CloosPAChristensenJAggerKMaiolicaARappsilberJ 2006 The putative oncogene GASC1 demethylates tri - and dimethylated lysine 9 on histone H3. Nature 442 307 311
24. LinJWuPHTarrPTLindenbergKSSt-PierreJ 2004 Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell 119 121 135
25. QiaoLMacDougaldOAShaoJ 2006 CCAAT/enhancer-binding protein alpha mediates induction of hepatic phosphoenolpyruvate carboxykinase by p38 mitogen-activated protein kinase. J Biol Chem 281 24390 24397
26. WangXLSuzukiRLeeKTranTGuntonJE 2009 Ablation of ARNT/HIF1beta in liver alters gluconeogenesis, lipogenic gene expression, and serum ketones. Cell Metab 9 428 439
27. QiaoLMacLeanPSYouHSchaackJShaoJ 2006 knocking down liver ccaat/enhancer-binding protein alpha by adenovirus-transduced silent interfering ribonucleic acid improves hepatic gluconeogenesis and lipid homeostasis in db/db mice. Endocrinology 147 3060 3069
28. LeeYHSauerBJohnsonPFGonzalezFJ 1997 Disruption of the c/ebp alpha gene in adult mouse liver. Mol Cell Biol 17 6014 6022
29. WangNDFinegoldMJBradleyAOuCNAbdelsayedSV 1995 Impaired energy homeostasis in C/EBP alpha knockout mice. Science 269 1108 1112
30. KimJWMonilaHPandeyALaneMD 2007 Upstream stimulatory factors regulate the C/EBP alpha gene during differentiation of 3T3-L1 preadipocytes. Biochem Biophys Res Commun 354 517 521
31. TadaYBrenaRMHackansonBMorrisonCOttersonGA 2006 Epigenetic modulation of tumor suppressor CCAAT/enhancer binding protein alpha activity in lung cancer. J Natl Cancer Inst 98 396 406
32. TateishiKOkadaYKallinEMZhangY 2009 Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 458 757 761
33. InagakiTTachibanaMMagooriKKudoHTanakaT 2009 Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice. Genes Cells 14 991 1001
34. XuLZhaoZDongASoubigou-TaconnatLRenouJP 2008 Di - and tri - but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol 28 1348 1360
35. CarrozzaMJLiBFlorensLSuganumaTSwansonSK 2005 Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123 581 592
36. JoshiAAStruhlK 2005 Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20 971 978
37. KeoghMCKurdistaniSKMorrisSAAhnSHPodolnyV 2005 Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123 593 605
38. HeJKallinEMTsukadaYZhangY 2008 The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat Struct Mol Biol 15 1169 1175
39. BlackledgeNPZhouJCTolstorukovMYFarcasAMParkPJ 2010 CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell 38 179 190
40. QiHHSarkissianMHuGQWangZBhattacharjeeA 2010 Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466 503 507
41. HallRKWangXLGeorgeLKochSRGrannerDK 2007 Insulin represses phosphoenolpyruvate carboxykinase gene transcription by causing the rapid disruption of an active transcription complex: a potential epigenetic effect. Mol Endocrinol 21 550 563
42. PanDFujimotoMLopesAWangYX 2009 Twist-1 is a PPARdelta-inducible, negative-feedback regulator of PGC-1alpha in brown fat metabolism. Cell 137 73 86
43. CampeauERuhlVERodierFSmithCLRahmbergBL 2009 A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE 4 e6529 doi:10.1371/journal.pone.0006529
44. HeTCZhouSda CostaLTYuJKinzlerKW 1998 A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A 95 2509 2514
45. AcevedoLGBiedaMGreenRFarnhamPJ 2008 Analysis of the mechanisms mediating tumor-specific changes in gene expression in human liver tumors. Cancer Res 68 2641 2651
Štítky
Genetika Reprodukční medicína
Článek Uracil-Containing DNA in : Stability, Stage-Specific Accumulation, and Developmental InvolvementČlánek Fuzzy Tandem Repeats Containing p53 Response Elements May Define Species-Specific p53 Target GenesČlánek Preferential Genome Targeting of the CBP Co-Activator by Rel and Smad Proteins in Early EmbryosČlánek Protective Coupling of Mitochondrial Function and Protein Synthesis via the eIF2α Kinase GCN-2Článek Cohesin Proteins Promote Ribosomal RNA Production and Protein Translation in Yeast and Human CellsČlánek TERRA Promotes Telomere Shortening through Exonuclease 1–Mediated Resection of Chromosome EndsČlánek Attenuation of Notch and Hedgehog Signaling Is Required for Fate Specification in the Spinal CordČlánek Genome-Wide Functional Profiling Identifies Genes and Processes Important for Zinc-Limited Growth ofČlánek MicroRNA93 Regulates Proliferation and Differentiation of Normal and Malignant Breast Stem Cells
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 6
-
Všechny články tohoto čísla
- Rumors of Its Disassembly Have Been Greatly Exaggerated: The Secret Life of the Synaptonemal Complex at the Centromeres
- Mimetic Butterflies Introgress to Impress
- Tipping the Balance in the Powerhouse of the Cell to “Protect” Colorectal Cancer
- Selection-Driven Gene Loss in Bacteria
- Decreased Mitochondrial DNA Mutagenesis in Human Colorectal Cancer
- Parallel Evolution of Auditory Genes for Echolocation in Bats and Toothed Whales
- Diverse CRISPRs Evolving in Human Microbiomes
- The Rad4 ATR-Activation Domain Functions in G1/S Phase in a Chromatin-Dependent Manner
- Stretching the Rules: Monocentric Chromosomes with Multiple Centromere Domains
- Uracil-Containing DNA in : Stability, Stage-Specific Accumulation, and Developmental Involvement
- Fuzzy Tandem Repeats Containing p53 Response Elements May Define Species-Specific p53 Target Genes
- Adaptive Introgression across Species Boundaries in Butterflies
- G Protein Activation without a GEF in the Plant Kingdom
- Synaptonemal Complex Components Persist at Centromeres and Are Required for Homologous Centromere Pairing in Mouse Spermatocytes
- An Engineering Approach to Extending Lifespan in
- Incompatibility and Competitive Exclusion of Genomic Segments between Sibling Species
- Effects of Histone H3 Depletion on Nucleosome Occupancy and Position in
- Patterns of Evolutionary Conservation of Essential Genes Correlate with Their Compensability
- Interplay between Synaptonemal Complex, Homologous Recombination, and Centromeres during Mammalian Meiosis
- Preferential Genome Targeting of the CBP Co-Activator by Rel and Smad Proteins in Early Embryos
- A Mouse Model of Acrodermatitis Enteropathica: Loss of Intestine Zinc Transporter ZIP4 (Slc39a4) Disrupts the Stem Cell Niche and Intestine Integrity
- Protective Coupling of Mitochondrial Function and Protein Synthesis via the eIF2α Kinase GCN-2
- Geographic Differences in Genetic Susceptibility to IgA Nephropathy: GWAS Replication Study and Geospatial Risk Analysis
- Cohesin Proteins Promote Ribosomal RNA Production and Protein Translation in Yeast and Human Cells
- TERRA Promotes Telomere Shortening through Exonuclease 1–Mediated Resection of Chromosome Ends
- Stimulation of Host Immune Defenses by a Small Molecule Protects from Bacterial Infection
- A Broad Requirement for TLS Polymerases η and κ, and Interacting Sumoylation and Nuclear Pore Proteins, in Lesion Bypass during Embryogenesis
- Genome-Wide Identification of Ampicillin Resistance Determinants in
- The CCR4-NOT Complex Is Implicated in the Viability of Aneuploid Yeasts
- Gustatory Perception and Fat Body Energy Metabolism Are Jointly Affected by Vitellogenin and Juvenile Hormone in Honey Bees
- Genetic Variants on Chromosome 1q41 Influence Ocular Axial Length and High Myopia
- Is a Key Regulator of Pancreaticobiliary Ductal System Development
- The NSL Complex Regulates Housekeeping Genes in
- Attenuation of Notch and Hedgehog Signaling Is Required for Fate Specification in the Spinal Cord
- Dual-Level Regulation of ACC Synthase Activity by MPK3/MPK6 Cascade and Its Downstream WRKY Transcription Factor during Ethylene Induction in Arabidopsis
- Genome-Wide Functional Profiling Identifies Genes and Processes Important for Zinc-Limited Growth of
- Base-Pair Resolution DNA Methylation Sequencing Reveals Profoundly Divergent Epigenetic Landscapes in Acute Myeloid Leukemia
- MicroRNA93 Regulates Proliferation and Differentiation of Normal and Malignant Breast Stem Cells
- Phylogenomic Analysis Reveals Dynamic Evolutionary History of the Drosophila Heterochromatin Protein 1 (HP1) Gene Family
- Found: The Elusive ANTAR Transcription Antiterminator
- The Mutation in Chickens Constitutes a Structural Rearrangement Causing Both Altered Comb Morphology and Defective Sperm Motility
- Control of CpG and Non-CpG DNA Methylation by DNA Methyltransferases
- Polymorphisms in the Mitochondrial Ribosome Recycling Factor Compromise Cell Respiratory Function and Increase Atorvastatin Toxicity
- Gene Expression Profiles in Parkinson Disease Prefrontal Cortex Implicate and Genes under Its Transcriptional Regulation
- Global Regulatory Functions of the Endoribonuclease III in Gene Expression
- Extensive Evolutionary Changes in Regulatory Element Activity during Human Origins Are Associated with Altered Gene Expression and Positive Selection
- The Regulatory Network of Natural Competence and Transformation of
- Brain Expression Genome-Wide Association Study (eGWAS) Identifies Human Disease-Associated Variants
- Quantifying the Adaptive Potential of an Antibiotic Resistance Enzyme
- Divergence of the Yeast Transcription Factor Affects Sulfite Resistance
- The Histone Demethylase Jhdm1a Regulates Hepatic Gluconeogenesis
- RNA Methylation by the MIS Complex Regulates a Cell Fate Decision in Yeast
- Rare Copy Number Variants Observed in Hereditary Breast Cancer Cases Disrupt Genes in Estrogen Signaling and Tumor Suppression Network
- Genome-Wide Location Analysis Reveals Distinct Transcriptional Circuitry by Paralogous Regulators Foxa1 and Foxa2
- A Mutation Links a Canine Progressive Early-Onset Cerebellar Ataxia to the Endoplasmic Reticulum–Associated Protein Degradation (ERAD) Machinery
- The Mechanism for RNA Recognition by ANTAR Regulators of Gene Expression
- Limits to the Rate of Adaptive Substitution in Sexual Populations
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Rumors of Its Disassembly Have Been Greatly Exaggerated: The Secret Life of the Synaptonemal Complex at the Centromeres
- The NSL Complex Regulates Housekeeping Genes in
- Tipping the Balance in the Powerhouse of the Cell to “Protect” Colorectal Cancer
- Interplay between Synaptonemal Complex, Homologous Recombination, and Centromeres during Mammalian Meiosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



