-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaGenome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
article has not abstract
Published in the journal: . PLoS Genet 8(11): e32767. doi:10.1371/journal.pgen.1003086
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1003086Summary
article has not abstract
DNA polymerase ß (Polß) is recognized as an essential DNA repair protein [1], [2]. Although the smallest of the human DNA polymerases [3], [4], this 335–amino-acid protein is the primary DNA polymerase in the base excision repair (BER) pathway [5]. A majority of the 20,000 DNA lesions per day that each human cell is faced with are repaired by the BER pathway [6]. These include products of base depurination and depyrimidination (abasic sites), deamination of cytosine and 5-methylcytosine, oxidation products such as 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG), thymine glycol and lipid peroxidation products, as well as methylation modifications such as N7-guanine [7], [8]. Failure to repair these spontaneous or endogenously induced DNA base lesions as well as the numerous base modifications that arise from environmental or exogenous sources can result in multiple cellular effects, including cell death, gene mutations, gene rearrangements, and/or decreased cell growth rate. Polß facilitates the repair of these base lesions in concert with different proteins of the BER pathway depending on the lesion [2], [7]. Once the base lesion is removed by one of 11 DNA glycosylase enzymes and the resulting abasic site is hydrolysed by the endonuclease APE1, Polß is recruited to the lesion via an interaction with the BER scaffold protein XRCC1 [9], [10] and the DNA damage sensor PARP1 [11]–[13]. Polß then conducts two essential enzymatic functions: 5′dRP lyase–mediated gap tailoring and DNA polymerase–mediated DNA synthesis to fill the gap [2], [3]. The 5′dRP lyase activity functions to “tailor” the gap by removing the sugar-phosphate residue that remains after APE1 cleaves the DNA backbone, and then the polymerase activity adds the newly synthesized nucleotide that was removed during repair. Considering the critical and essential role of these two enzymatic activities, the important protein–protein interactions between Polß and several BER proteins [14], as well as the increasing number of post-translational modifications suggested to affect Polß function and stability [15], it may not be surprising that a significant number of somatic mutations in POLB have been observed in cancer (Table 1). Within the 33 Kb POLB gene (PubMed geneID #5423), as many as 567 SNPs have been identified (see dbSNP). However, only 34 SNPs are in or near the coding region (22 are found in exons), and only two have been confirmed in larger cohorts. These two germline POLB mutants (R137Q; rs12678588 and P242R; rs3136797) have been reported to be present in as much as 0.6% and 2.4% of the human population, respectively [16], [17]. However, little is known about the functional impact that results from these single amino acid alterations. An earlier study on the Polß (R137Q) mutant (rs12678588) suggested that the R137Q mutation impairs function of the purified protein. Further, when produced in mouse cells, the R137Q mutant protein interfered with Polß binding to PCNA [18] and the response of mouse cells to DNA-damaging agents, although no information was provided on the impact of this mutation on genome stability. Whereas the Polß (P242R) mutant allele (rs3136797) has been linked with altered incidence of cancer in several studies [19]–[21], there have been few or no studies defining the impact of this SNP on Polß function, DNA repair capacity, and genome maintenance in human cells.
Tab. 1. Germline and somatic POLB mutants.* 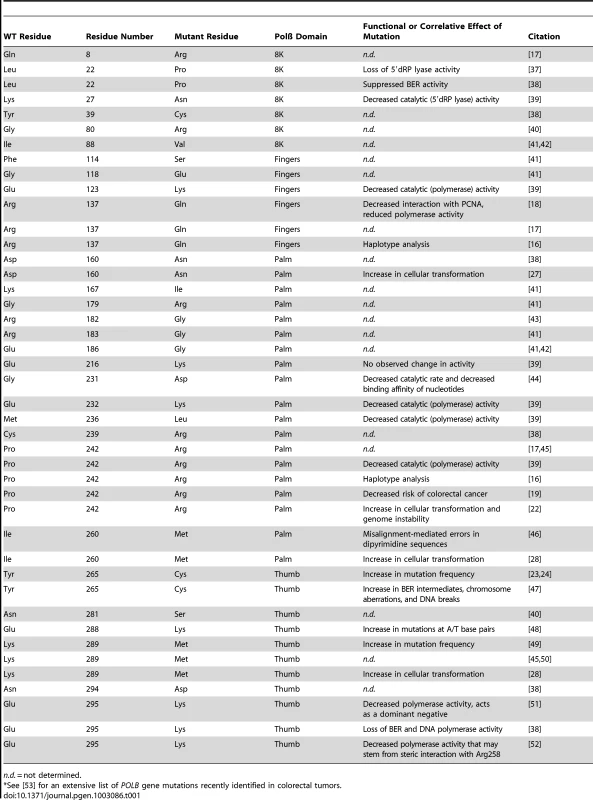
n.d. = not determined. In this issue of PLOS Genetics, Sweasy and colleagues conducted a detailed analysis of the POLB germline–coding SNP rs3136797 [22]. This polymorphism alters amino acid 242, changing the amino acid from a proline (P; Pro) to an arginine (R; Arg). To determine whether the P242R mutation affected genome stability in human or mouse cells, the wild-type (WT) or P242R protein was produced in human normal mammary epithelial cells (MCF10A) and in mouse embryonic fibroblasts (MEFs). In both cell lines, the synthesis of the P242R protein led to an increase in genomic alterations. Analysis of metaphase spreads showed that the P242R protein induced an increase in chromosome breaks and a significant increase in fragmented chromosomes and chromosome fusions. In other reports, cancer-specific mutations in Polß (e.g., Y265C) induced an increase in mutant frequency [23], [24] that could explain the increase in chromosome alterations seen with P242R. However, cells producing the P242R protein were found to have the same mutant frequency as those expressing WT Polß. The lack of an increase in mutations together with the increase in chromosomal instability suggested that the Polß (P242R) protein may promote the accumulation of DNA strand breaks during repair. To test this hypothesis, the cells were treated with methyl methanesulfonate (MMS) to induce DNA damage repaired by Polß [5]. As suspected, exposure of the cells expressing the P242R mutant to MMS induced a greater level of single-strand and double-strand DNA breaks. The increase in single-strand breaks and related BER intermediates was measured by the alkaline Comet assay [25], and an increase in the number of DNA double-strand breaks was indirectly determined by measuring an increase in γ-H2AX foci [26]. A second phenotype that Sweasy and colleagues have linked with cancer mutants of Polß is the ability to induce cellular transformation, as was seen with the D160N, I260M, and K289M Polß mutants [27], [28]. Similarly, production of the P242R mutant protein in mouse cells (C127λ) or human cells (MCF10A) increased growth in soft agar significantly in comparison with expression of WT Polß.
Yamtich et al. [22] then used both cellular analysis and biochemical measurements to evaluate the functional impact of the P242R mutation. WT MEF cells (expressing endogenous WT Polß) were engineered to produce either WT human Polß or the P242R mutant protein and were then exposed to MMS to measure cellular survival. Both WT - and P242R-expressing cells responded equally except at the highest doses of MMS. Next, Polß-knock out (KO) MEFs engineered to express either WT human Polß or the P242R mutant were exposed to MMS to determine whether the P242R mutant could restore (complement) resistance to MMS. In this case, there was a small but significant difference in response, suggesting that the P242R mutant was mildly defective in BER. A strength of the Sweasy lab's study is the use of cell biology analyses as well as detailed biochemical evaluation of these mutant proteins. Yamtich et al. [22] expressed and purified the WT and P242R mutant proteins from E. coli and measured the rate of DNA polymerase activity by two separate kinetic analyses. This provided the opportunity to determine whether the decreased BER capacity observed in the Polß-KO MEFs expressing the P242R mutant in response to MMS and the increase in DNA breaks were the result of a defect in the polymerase activity of the P242R mutant. In both cases, they found that the mutation (P242R) caused a decrease in the rate of DNA polymerase activity. However, the protein bound to the DNA substrate with affinity equal to that of the WT enzyme. The slow polymerase activity of the P242R protein therefore is likely to promote the accumulation of BER intermediates, inducing genome alterations when the cell is exposed to DNA-damaging agents [29].
Defects in Polß can have significant cellular ramifications, especially in response to DNA-damaging agents that require Polß and BER for repair. Complete loss of Polß function can trigger an increase in cell death in response to high doses of genotoxins [5], [30] and an increase in genome alterations even at low doses [29]. Additional cellular responses to DNA damage when Polß is defective may include PARP1 activation and alterations in bioenergetic metabolites such as NAD+ [31]. The steady-state expression level of Polß is also reported to be regulated by the proteasome via ubiquitylation [32], suggesting that some Polß mutants may have altered stability. In this regard, the observation that the P242R mutant protein has a functional defect now opens the door for further studies to clarify the mechanisms and cellular impacts of other defects in Polß. It has been suggested that tumor-specific defects in BER, such as a defect in Polß, may be exploited for selective therapeutic options [33]. Cells producing the mutant protein (P242R) have a higher level of DNA strand breaks and increased cellular transformation, and so it is possible that the Polß (P242R) protein may be considered a driver of cancer formation. It remains to be determined whether the presence of this mutant protein (P242R) provides therapeutic selectivity.
Finally, it remains to be determined how a mutation (P242R) so distant from the Polß active site, and which does not interfere with binding to XRCC1 (Figure 1), can have such a significant effect on the function of Polß. Given the subtle yet significant impact of the Polß (P242R) mutant on cellular function and genome stability in response to DNA damage as described by Yamtich et al [22], further analysis of this mutant protein is warranted. The P242 amino acid is located in a loop domain that is essential for enzymatic activity [34], so it is likely that the alteration of the amino acid from P to R changes the movement of the loop and may also change the overall architecture of the protein. To more completely appreciate the subtle yet significant defect associated with this germline mutation, it is therefore suggested that future studies be conducted to determine the structure of the ternary complex of Polß (P242R) with DNA and an incoming nucleotide. In addition, whole animal studies should be considered so as to determine whether the genome instability and cellular transformation results described [22] extend to additional cell types. As a germline mutation, analysis of the P242R mutant protein in an animal model will provide valuable insight into the possible effects on human health.
Fig. 1. Model depicting the structure of Polß. 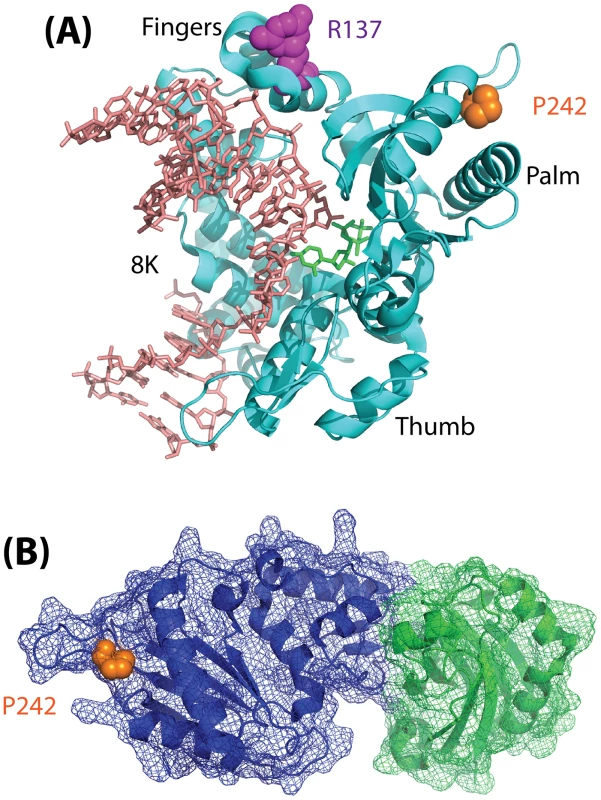
(A) Structure (pdb2fms) depicting DNA Polymerase ß (Polß) with a gapped DNA substrate and dUMPNPP with magnesium in the catalytic site [35]. The image is a cartoon rendition of the polypeptide chain of Polß in teal, the gapped DNA substrate in salmon, and the incoming dUMPNPP base in green. Amino acids known to be altered by germline mutations are shown using a space-filling rendering: R137 (magenta) and P242 (orange). The fingers, palm, and thumb domains of Polß are indicated. The 8K domain is at the back of the structure, facing away from the plane of the image, and is shown behind the DNA in this orientation. (B) Structure (pdb3lqc) depicting oxidized XRCC1 bound to the Polß palm/thumb domains [36]. The image is a cartoon rendition of the palm and thumb domains of Polß in blue, with a mesh illustrating the surface of the structure (amino acids 150–335), and a cartoon rendition of the oxidized form of XRCC1 in green, with a mesh illustrating the surface of the structure (amino acids 1–151). Amino acid P242 (orange) is shown using a space-filling rendering. The images were generated using PyMOL (Molecular Graphics System, Version 1.2r3pre; Schrödinger, LLC; http://pymol.org/).
Zdroje
1. BurgersPM, KooninEV, BrufordE, BlancoL, BurtisKC, et al. (2001) Eukaryotic DNA polymerases: proposal for a revised nomenclature. J Biol Chem 276 : 43487–43490.
2. AlmeidaKH, SobolRW (2007) A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair 6 : 695–711.
3. BeardWA, WilsonSH (2006) Structure and mechanism of DNA polymerase Beta. Chem Rev 106 : 361–382.
4. LangeSS, TakataK, WoodRD (2011) DNA polymerases and cancer. Nature Reviews Cancer 11 : 96–110.
5. SobolRW, HortonJK, KuhnR, GuH, SinghalRK, et al. (1996) Requirement of mammalian DNA polymerase-ß in base-excision repair. Nature 379 : 183–186.
6. LindahlT (1993) Instability and decay of the primary structure of DNA. Nature 362 : 709–715.
7. SvilarD, GoellnerEM, AlmeidaKH, SobolRW (2011) Base excision repair and lesion-dependent sub-pathways for repair of oxidative DNA damage. Antioxid Redox Signal 14 : 2491–2507.
8. Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, et al.. (2006) DNA repair and mutagenesis, 2nd edition. Washington, D.C.: ASM Press. 1,164 p.
9. CaldecottKW, AoufouchiS, JohnsonP, ShallS (1996) XRCC1 polypeptide interacts with DNA polymerase ß and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Research 24 : 4387–4394.
10. LanL, NakajimaS, OohataY, TakaoM, OkanoS, et al. (2004) In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proceedings of the National Academy of Science 101 : 13738–13743.
11. JelezcovaE, TrivediRN, WangXH, TangJB, BrownAR, et al. (2010) Parp1 activation in mouse embryonic fibroblasts promotes Pol beta-dependent cellular hypersensitivity to alkylation damage. Mutat Res 686 : 57–67.
12. TangJB, SvilarD, TrivediRN, WangXH, GoellnerEM, et al. (2011) N-methylpurine DNA glycosylase and DNA polymerase beta modulate BER inhibitor potentiation of glioma cells to temozolomide. Neuro Oncol 13 : 471–486.
13. MasaokaA, HortonJK, BeardWA, WilsonSH (2009) DNA polymerase beta and PARP activities in base excision repair in living cells. DNA Repair (Amst) 8 : 1290–1299.
14. Almeida KH, Sobol RW (2005) Increased specificity and efficiency of base excision repair through complex formation. In: Siede W, Doetsch PW, Kow YW, editors. DNA damage recognition. New York: Marcel Dekker Inc. pp. 33–64.
15. SobolRW (2008) CHIPping away at base excision repair. Molecular Cell 29 : 413–415.
16. YamtichJ, SpeedWC, StrakaE, KiddJR, SweasyJB, et al. (2009) Population-specific variation in haplotype composition and heterozygosity at the POLB locus. DNA Repair (Amst) 8 : 579–584.
17. MohrenweiserHW, XiT, Vazquez-MatiasJ, JonesIM (2002) Identification of 127 amino acid substitution variants in screening 37 DNA repair genes in humans. Cancer Epidemiology Biomarkers & Prevention 11 : 1054–1064.
18. GuoZ, ZhengL, DaiH, ZhouM, XuH, et al. (2009) Human DNA polymerase beta polymorphism, Arg137Gln, impairs its polymerase activity and interaction with PCNA and the cellular base excision repair capacity. Nucleic Acids Res 37 : 3431–3441.
19. MorenoV, GemignaniF, LandiS, Gioia-PatricolaL, ChabrierA, et al. (2006) Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res 12 : 2101–2108.
20. MatakidouA, el GaltaR, WebbEL, RuddMF, BridleH, et al. (2007) Genetic variation in the DNA repair genes is predictive of outcome in lung cancer. Hum Mol Genet 16 : 2333–2340.
21. SellickGS, WadeR, RichardsS, OscierDG, CatovskyD, et al. (2008) Scan of 977 nonsynonymous SNPs in CLL4 trial patients for the identification of genetic variants influencing prognosis. Blood 111 : 1625–1633.
22. YamtichJ, NemecAA, KehA, SweasyJB (2012) A germline polymorphism of DNA polymerase beta induces genomic instability and cellular transformation. PLoS Genet 8: e1003052 doi:10.1371/journal.pgen.1003052
23. ClairmontCA, NarayananL, SunKW, GlazerPM, SweasyJB (1999) The Tyr-265-to-Cys mutator mutant of DNA polymerase beta induces a mutator phenotype in mouse LN12 cells. Proceedings of the National Acadamy of Sciences USA 96 : 9580–9585.
24. OpreskoPL, SweasyJB, EckertKA (1998) The mutator form of polymerase beta with amino acid substitution at tyrosine 265 in the hinge region displays an increase in both base substitution and frame shift errors. Biochemistry 37 : 2111–2119.
25. FortiniP, RaspaglioG, FalchiM, DogliottiE (1996) Analysis of DNA alkylation damage and repair in mammalian cells by the comet assay. Mutagenesis 11 : 169–175.
26. PaullTT, RogakouEP, YamazakiV, KirchgessnerCU, GellertM, et al. (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Current Biology 10 : 886–895.
27. DoniganKA, HileSE, EckertKA, SweasyJB (2012) The human gastric cancer-associated DNA polymerase beta variant D160N is a mutator that induces cellular transformation. DNA Repair (Amst) 11 : 381–390.
28. SweasyJB, LangT, StarcevicD, SunKW, LaiCC, et al. (2005) Expression of DNA polymerase {beta} cancer-associated variants in mouse cells results in cellular transformation. Proc Natl Acad Sci U S A 102 : 14350–14355.
29. SobolRW, KartalouM, AlmeidaKH, JoyceDF, EngelwardBP, et al. (2003) Base excision repair intermediates induce p53-independent cytotoxic and genotoxic responses. Journal of Biological Chemistry 278 : 39951–39959.
30. SobolRW, PrasadR, EvenskiA, BakerA, YangXP, et al. (2000) The lyase activity of the DNA repair protein ß-polymerase protects from DNA-damage-induced cytotoxicity. Nature 405 : 807–810.
31. TangJ, GoellnerEM, WangXW, TrivediRN, St. CroixCM, et al. (2010) Bioenergetic metabolites regulate base excision repair-dependent cell death in response to DNA damage. Molecular Cancer Research 8 : 67–79.
32. ParsonsJL, TaitPS, FinchD, DianovaII, AllinsonSL, et al. (2008) CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol Cell 29 : 477–487.
33. NeijenhuisS, Verwijs-JanssenM, van den BroekLJ, BeggAC, VensC (2010) Targeted radiosensitization of cells expressing truncated DNA polymerase {beta}. Cancer Research 70 : 8706–8714.
34. LinGC, JaegerJ, EckertKA, SweasyJB (2009) Loop II of DNA polymerase beta is important for discrimination during substrate binding. DNA Repair (Amst) 8 : 182–189.
35. BatraVK, BeardWA, ShockDD, KrahnJM, PedersenLC, et al. (2006) Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure 14 : 757–766.
36. CuneoMJ, LondonRE (2010) Oxidation state of the XRCC1 N-terminal domain regulates DNA polymerase beta binding affinity. Proceedings of the National Acadamy of Sciences USA 107 : 6805–6810.
37. DalalS, ChikovaA, JaegerJ, SweasyJB (2008) The Leu22Pro tumor-associated variant of DNA polymerase beta is dRP lyase deficient. Nucleic Acids Res 36 : 411–422.
38. IwanagaA, OuchidaM, MiyazakiK, HoriK, MukaiT (1999) Functional mutation of DNA polymerase ß found in human gastric cancer: inability of the base excision repair in vitro. Mutation Research 435 : 121–128.
39. AnCL, ChenD, MakridakisNM (2011) Systematic biochemical analysis of somatic missense mutations in DNA polymerase beta found in prostate cancer reveal alteration of enzymatic function. Hum Mutat 32 : 415–423.
40. ForbesSA, BindalN, BamfordS, ColeC, KokCY, et al. (2011) COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 39: D945–950.
41. DongZ, ZhaoG, ZhaoQ, YangH, XueL, et al. (2002) [A study of DNA polymerase beta mutation in human esophageal cancer]. Zhonghua Yi Xue Za Zhi 82 : 899–902.
42. ZhaoGQ, WangT, ZhaoQ, YangHY, TanXH, et al. (2005) Mutation of DNA polymerase beta in esophageal carcinoma of different regions. World J Gastroenterol 11 : 4618–4622.
43. HanLP, QiaoYH, DongZM, ShiHR, ZhaoGQ, et al. (2003) [Study on DNA polymerase beta gene mutation in human cervical cancer]. Zhonghua Fu Chan Ke Za Zhi 38 : 618–620.
44. NemecAA, DoniganKA, MurphyDL, JaegerJ, SweasyJB (2012) Colon cancer-associated DNA polymerase beta variant induces genomic instability and cellular transformation. Journal of Biological Chemistry 287 : 23840–23849.
45. SliwinskiT, ZiembaP, MorawiecZ, KowalskiM, ZadroznyM, et al. (2007) Polymorphisms of the DNA polymerase beta gene in breast cancer. Breast Cancer Res Treat 103 : 161–166.
46. DalalS, HileS, EckertKA, SunKW, StarcevicD, et al. (2005) Prostate-cancer-associated I260M variant of DNA polymerase beta is a sequence-specific mutator. Biochemistry 44 : 15664–15673.
47. SenejaniAG, DalalS, LiuY, NottoliTP, McGrathJM, et al. (2012) Y265C DNA polymerase beta knockin mice survive past birth and accumulate base excision repair intermediate substrates. Proceedings of the National Acadamy of Sciences USA 109 : 6632–6637.
48. MurphyDL, DoniganKA, JaegerJ, SweasyJB (2012) The E288K colon tumor variant of DNA polymerase beta is a sequence specific mutator. Biochemistry 51 : 5269–5275.
49. LangT, MaitraM, StarcevicD, LiSX, SweasyJB (2004) A DNA polymerase beta mutant from colon cancer cells induces mutations. Proc Natl Acad Sci U S A 101 : 6074–6079.
50. WangL, PatelU, GhoshL, BanerjeeS (1992) DNA polymerase ß mutations in human colorectal cancer. Cancer Research 52 : 4824–4827.
51. LangT, DalalS, ChikovaA, DimaioD, SweasyJB (2007) The E295K DNA polymerase beta gastric cancer-associated variant interferes with base excision repair and induces cellular transformation. Mol Cell Biol 27 : 5587–5596.
52. LiY, GridleyCL, JaegerJ, SweasyJB, SchlickT (2012) Unfavorable electrostatic and steric interactions in DNA polymerase beta E295K mutant interfere with the enzyme's pathway. J Am Chem Soc 134 : 9999–10010.
53. DoniganKA, SunKW, NemecAA, MurphyDL, CongX, et al. (2012) Human POLB gene is mutated in high percentage of colorectal tumors. Journal of Biological Chemistry 287 : 23830–23839.
Štítky
Genetika Reprodukční medicína
Článek The Covariate's DilemmaČlánek Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene SignallingČlánek Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 11
-
Všechny články tohoto čísla
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- The Covariate's Dilemma
- Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
- Plan B for Stimulating Stem Cell Division
- Discovering Thiamine Transporters as Targets of Chloroquine Using a Novel Functional Genomics Strategy
- Is a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
- Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
- Genome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
- Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant
- Controls of Nucleosome Positioning in the Human Genome
- Disruption of Causes Defective Meiotic Recombination in Male Mice
- A Novel Human-Infection-Derived Bacterium Provides Insights into the Evolutionary Origins of Mutualistic Insect–Bacterial Symbioses
- Trps1 and Its Target Gene Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Population-Based Resequencing of in 10,330 Individuals: Spectrum of Genetic Variation, Phenotype, and Comparison with Extreme Phenotype Approach
- HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in
- Transcription Elongation and Tissue-Specific Somatic CAG Instability
- A Germline Polymorphism of DNA Polymerase Beta Induces Genomic Instability and Cellular Transformation
- Interallelic and Intergenic Incompatibilities of the () Gene in Mouse Hybrid Sterility
- Comparison of Mitochondrial Mutation Spectra in Ageing Human Colonic Epithelium and Disease: Absence of Evidence for Purifying Selection in Somatic Mitochondrial DNA Point Mutations
- Mutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
- Evolution of Minimal Specificity and Promiscuity in Steroid Hormone Receptors
- Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
- RHOA Is a Modulator of the Cholesterol-Lowering Effects of Statin
- MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways
- Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in
- Genome-Wide Association Studies Reveal a Simple Genetic Basis of Resistance to Naturally Coevolving Viruses in
- The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the Class I Antigen Recognition Groove
- Molecular, Physiological, and Motor Performance Defects in DMSXL Mice Carrying >1,000 CTG Repeats from the Human DM1 Locus
- Genomic Study of RNA Polymerase II and III SNAP-Bound Promoters Reveals a Gene Transcribed by Both Enzymes and a Broad Use of Common Activators
- Long Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
- The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing
- Deubiquitylation Machinery Is Required for Embryonic Polarity in
- dJun and Vri/dNFIL3 Are Major Regulators of Cardiac Aging in Drosophila
- CtIP Is Required to Initiate Replication-Dependent Interstrand Crosslink Repair
- Notch-Mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Intestinal Stem Cell Lineage
- A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation
- Network Analysis of a -Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier
- Mitosis in Neurons: Roughex and APC/C Maintain Cell Cycle Exit to Prevent Cytokinetic and Axonal Defects in Photoreceptor Neurons
- CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function
- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- The Genomes of the Fungal Plant Pathogens and Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry
- A Genome-Scale RNA–Interference Screen Identifies RRAS Signaling as a Pathologic Feature of Huntington's Disease
- Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
- Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites
- Tissue-Specific Regulation of Chromatin Insulator Function
- Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome
- Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga CCMP1779
- Antagonistic Gene Activities Determine the Formation of Pattern Elements along the Mediolateral Axis of the Fruit
- Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma
- Identification of the First ATRIP–Deficient Patient and Novel Mutations in ATR Define a Clinical Spectrum for ATR–ATRIP Seckel Syndrome
- Cooperativity of , , and in Malignant Breast Cancer Evolution
- Loss of Prohibitin Membrane Scaffolds Impairs Mitochondrial Architecture and Leads to Tau Hyperphosphorylation and Neurodegeneration
- Microhomology Directs Diverse DNA Break Repair Pathways and Chromosomal Translocations
- MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
- Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme
- The Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
- Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
- Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
- A Unified Method for Detecting Secondary Trait Associations with Rare Variants: Application to Sequence Data
- Genetic and Biochemical Dissection of a HisKA Domain Identifies Residues Required Exclusively for Kinase and Phosphatase Activities
- Informed Conditioning on Clinical Covariates Increases Power in Case-Control Association Studies
- Biochemical Diversification through Foreign Gene Expression in Bdelloid Rotifers
- Genomic Variation and Its Impact on Gene Expression in
- Spastic Paraplegia Mutation N256S in the Neuronal Microtubule Motor KIF5A Disrupts Axonal Transport in a HSP Model
- Lamin B1 Polymorphism Influences Morphology of the Nuclear Envelope, Cell Cycle Progression, and Risk of Neural Tube Defects in Mice
- A Targeted Glycan-Related Gene Screen Reveals Heparan Sulfate Proteoglycan Sulfation Regulates WNT and BMP Trans-Synaptic Signaling
- Dopaminergic D2-Like Receptors Delimit Recurrent Cholinergic-Mediated Motor Programs during a Goal-Oriented Behavior
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



