-
Články
Reklama
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
ReklamaKDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
Reactive oxygen species are essential signaling molecules within the cell. However, when levels of these reactive intermediates become too high (oxidative stress), they cause significant damage to proteins and DNA. It is therefore vitally important to understand how cells regulate genes required to limit oxidative stress. Here we describe a new role for the transcription co-factor KDM5 as an activator of genes that prevent oxidative stress. KDM5 activates these genes by interacting with the transcription factor Foxo and affecting its ability to be recruited to target promoters. Our data provide new insights into the mechanisms by which redox state is regulated, and into the multiple means by which KDM5 regulates gene expression.
Published in the journal: . PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004676
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004676Summary
Reactive oxygen species are essential signaling molecules within the cell. However, when levels of these reactive intermediates become too high (oxidative stress), they cause significant damage to proteins and DNA. It is therefore vitally important to understand how cells regulate genes required to limit oxidative stress. Here we describe a new role for the transcription co-factor KDM5 as an activator of genes that prevent oxidative stress. KDM5 activates these genes by interacting with the transcription factor Foxo and affecting its ability to be recruited to target promoters. Our data provide new insights into the mechanisms by which redox state is regulated, and into the multiple means by which KDM5 regulates gene expression.
Introduction
Tightly regulated transcription is essential for developmental and homeostatic processes, and is an important means by which animals respond to environmental cues. In addition to sequence-specific transcription factors that activate or repress transcription, an additional layer of gene expression regulation is provided by covalent modifications that occur on the tails of nucleosomal histones such as methylation and acetylation [1]. These chromatin modifications can alter DNA compaction to influence the ability of transcription factors to bind, and form docking sites for additional regulatory proteins that affect gene expression [2]. KDM5 proteins are an important family of transcriptional co-factors because they can both recognize and enzymatically modify specific chromatin modifications [3], [4]. KDM5 proteins are therefore able to regulate gene expression by more than one mechanism leading to context-dependent activation or repression of transcription.
Utilizing their well-characterized Jumonji (JmjC) domain, KDM5 proteins function as lysine demethylases [3]–[5]. The four mammalian KDM5 paralogs can demethylate di - and trimethylated histone H3 lysine 4 (H3K4me2/3) while the sole Drosophila KDM5 ortholog specifically targets H3K4me3 [6]–[13]. Because H3K4me2/3-containing nucleosomes are primarily found at promoter regions and are a hallmark of actively transcribed genes, removal of these chromatin marks by KDM5 demethylases correlates with transcriptional repression [14]. Interestingly, while Drosophila KDM5 and mouse KDM5B are essential genes, the demethylase function of these proteins is dispensable for viability [15]–[17]. This finding highlights the importance of the other gene-regulatory activities of KDM5 proteins. Indeed, KDM5 family proteins can influence gene expression through changes to histone acetylation by interacting with lysine deacetylases such as HDAC1 and HDAC4 [18]–[20]. In addition, KDM5 proteins can bind to unmodified histone H3 and histone H3 that is methylated on lysine 4 via their PHD finger motifs, although the biological importance of these chromatin-recognition activities remains largely uncharacterized [15], [21].
Emphasizing the importance of their gene-regulatory functions, fly KDM5 and mouse KDM5B are essential for viability [17], [22]. Moreover, three of the four human paralogs are implicated in disease. KDM5A and KDM5B are overexpressed in a large number of tumors including melanoma, breast, gastric and lung cancer (reviewed by [3]). While the precise role of KDM5A and KDM5B in tumor formation remains to be elucidated, recent evidence supports the interesting notion that KDM5B overexpression promotes the growth and survival of cancer “stem cells” [23], [24]. These oncogenic functions may be explained in part by the observation made by us and others that KDM5 family proteins interact with the well-known oncoprotein Myc and the tumor suppressor pRB [13], [15], [25]–[28]. Unlike dysregulation of KDM5A and KDM5B that are linked to cancer, mutations in KDM5C are found in patients with syndromic and non-syndromic intellectual disability [29]–[32]. This suggests that this family member plays an essential role in neuronal development. Consistent with this, KDM5C is expressed at high levels in neuronal cells and its knockdown in cultured rat cerebellar neurons causes dendritic defects [8]. KDM5 family proteins are therefore of central importance for normal cellular growth and function in a range of cell types.
Here we describe KDM5 as a new regulator of cellular levels of oxidative stress. Moreover, this occurs by a previously undescribed mechanism in which KDM5 influences transcription factor binding. Our data demonstrate that Drosophila KDM5 directly activates genes required to regulate cellular redox state. Consistent with this, kdm5 mutants are sensitive to treatment with the oxidizer paraquat, show elevated levels of oxidized proteins, and have an increased mutation load. Significantly, KDM5 interacts with the well-established oxidative stress transcription factor Foxo and is required for its recruitment to target promoters. These data expand the repertoire of KDM5's gene-regulatory functions and significantly add to our understanding of how transcription factors find and bind to their cognate DNA binding sites in vivo. Because increased cellular oxidative stress is implicated in cancer and neuronal dysfunction [33]–[35], our data have clear implications for understanding how dysregulation of KDM5 family proteins results in human disease.
Results
kdm5 mutants have deregulated expression of genes required for oxidative stress resistance
To determine the gene expression changes associated with loss of KDM5, we carried out microarray analyses of dissected wing imaginal discs from wildtype (w1118) and kdm510424 homozygous mutant 3rd instar larvae. kdm510424 behaves genetically as a strong loss of function allele and we are unable to detect KDM5 protein by Western blot (Figure 1A). These analyses revealed that of the 18,500 transcripts represented on the array, 367 genes were up-regulated and 534 genes were down-regulated 1.5-fold or more in kdm5 mutants compared to wildtype (p<0.01; Figure 1B; Figure S1A–F). A majority of genes up or down-regulated in kdm5 mutant wing discs were moderately affected (1.5 to 5-fold), suggesting that KDM5 does not cause dramatic changes to gene expression levels (Figure S1G). Using real time-PCR we confirmed the expression of five downregulated and five upregulated transcripts, validating our microarray data (Figure S1H).
Fig. 1. KDM5 mutants show reduced expression of genes required to reduce oxidative stress. 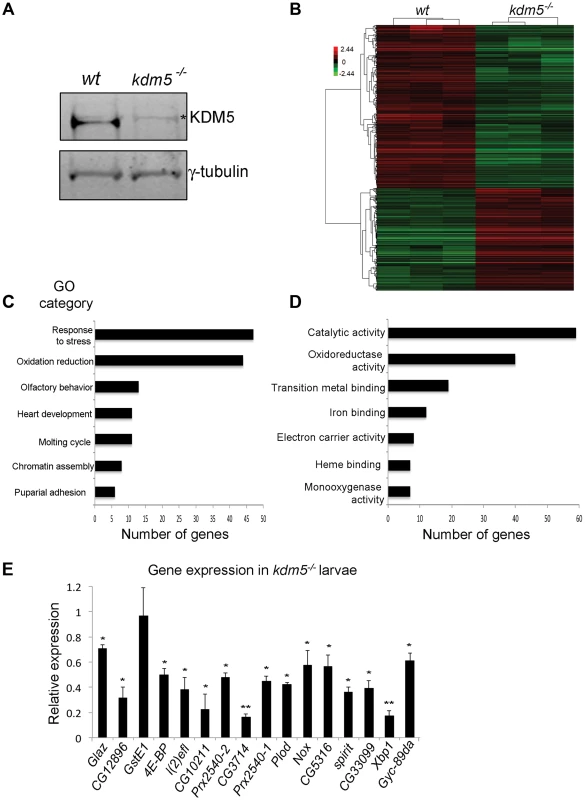
A) Western blot showing levels of KDM5 and the loading control γ-tubulin in wildtype (w1118) and kdm510424 homozygous mutant wing imaginal discs used for microarrays. * indicates a non-specific band. (B) Hierarchical clustering analysis of 901 genes that are differentially expressed 1.5-fold or more in kdm5 mutant wing discs compared to wildtype (w1118) (p<0.05). Up-regulated genes are shown in red; down-regulated genes in green. (C) Gene ontology analyses showing biological process enrichment analysis of down- and up-regulated genes (p≤0.01). (D) GO David analyses of the Molecular function of the 84 stress response and oxidation reduction class genes (p≤0.01). (E) Real-time PCR showing levels of oxidative stress response genes in kdm510424/K6801 3rd instar female larvae relative to wildtype. Levels of gene expression within each sample were normalized to rp49 and shown relative to wildtype. * p<0.05, ** p<0.01. To highlight biological processes regulated by KDM5, we determined which gene ontology (GO) terms were enriched within the differentially expressed genes (both up and downregulated). While a total of ten GO terms were identified, two functionally related and overlapping terms had the largest number of genes: (1) response to stress and (2) oxidation reduction (Figure 1C). These two classes had 45 and 44 genes, respectively, with 7 that overlap. 45 of these 82 stress response and oxidation reduction genes were downregulated and 37 were upregulated (Figure 1D; Figure S2). Downregulated genes include those with established roles in responding to oxidative and other forms of stress. For example the translation initiation factor 4E-BP that, while also involved in other processes, also regulates the translation of components of mitochondrial complex I and is required for survival in conditions of oxidative stress [36]–[38]. In addition, kdm5 mutants show downregulation of peroxiredoxins that detoxify H2O2 (e.g. Prx2540-1, Prx2540-2, CG12896, CG10211) [39]–[41], maintain cellular NAD levels (CG3714) [42], or function as chaperones (Glaz, l(2)efl) [43], [44]. In contrast, upregulated genes include methuselah (mth) family genes that negatively regulate oxidative stress resistance [45]. These data suggest that KDM5 may be a new regulator of cellular redox state.
To understand the role of KDM5 in the transcriptional regulation of oxidative stress resistance, we focused on 16 genes involved in this process that were downregulated in our kdm5 mutant microarray data. These genes require KDM5 for their activation, and the mechanisms by which KDM5 induces gene expression are not well characterized. To examine their expression in whole 3rd instar larvae we used animals homozygous for kdm510424 or transheterozygous for the allelic combination of kdm510424 and kdm5K06801 (kdm510424/K06801). All kdm5 mutants examined showed reduced expression of 15 out of the 16 genes tested (Figure 1E). While one gene (GstE1) was significantly downregulated in wing imaginal discs, it was not reduced in whole larvae, suggesting that its regulation by KDM5 is tissue specific. KDM5 is therefore required to maintain the expression of oxidative stress resistance genes in many other larval tissues in addition to wing disc cells where this regulation was first identified.
KDM5 is required for survival in conditions of oxidative stress
Because mutations in several genes that require KDM5 for their activation show sensitivity to conditions of oxidative stress (e.g. 4E-BP, Glaz, l(2)efl; [37], ), we tested whether kdm5 mutant larvae were sensitive to treatment with the established oxidizer paraquat. While kdm5 mutant larvae survive in a similar manner to wildtype in non-stressed conditions, they die at a significantly higher rate when exposed to paraquat for six hours (Figure 2A). KDM5 is therefore required for larval survival to conditions of acute oxidative stress.
Fig. 2. Reducing KDM5 affects cellular levels of oxidative stress. 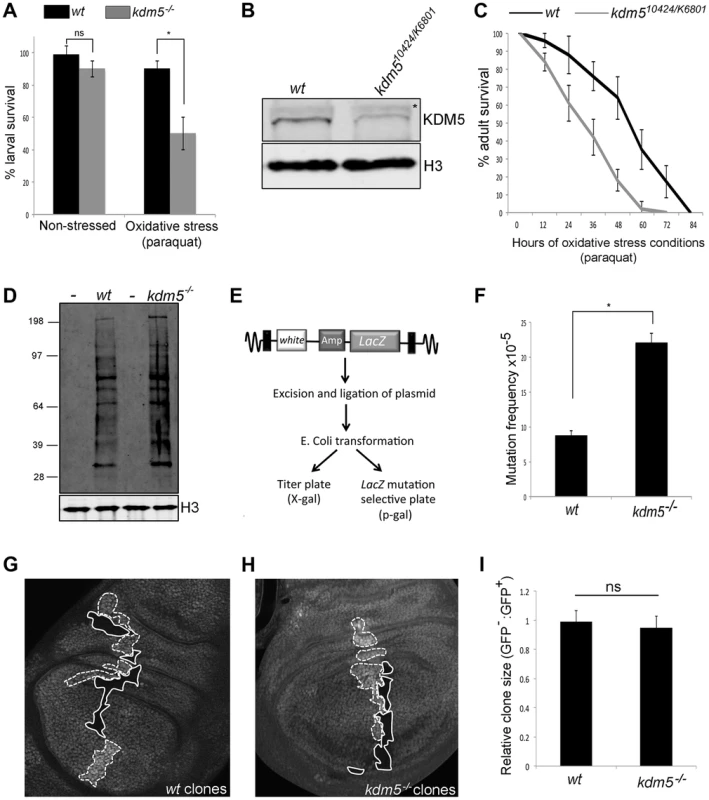
(A) Survival of wildtype control (w1118) and kdm510424/K06801 female larvae in 5% sucrose (left) or in 20 mM paraquat/5% sucrose (right) for six hours. * p<0.01. (B) Western blot showing levels of KDM5 and the loading control histone H3 in wildtype (w1118) and kdm510424/K6801 adult female heads (three heads per lane). (C) Survival curve of w1118 and kdm510424/K06801 adult females fed 20 mM paraquat in 5% sucrose. Adults were one to three days old at the start of the experiment. Error bars represent standard error. The two survival curves are significantly different from one another (p<0.05). (D) Detection of oxidized proteins in wildtype and kdm5K6801 homozygous mutant 3rd instar larvae using oxyblot. Molecular weight markers are shown on the left. – indicates negative control lanes in which carbonyl groups were not derivatized. kdm5 mutants show ∼2-fold increase in total levels of oxidized proteins. Histone H3 is shown as a loading control. (E) Schematic of lacZ mutation reporter assay. Adapted from Garcia et al [51]. (F) lacZ mutation frequency of wildtype control (w1118) and kdm510424 homozygous mutant 3rd instar larvae. Genotypes are w1118; #9lacZ; + (labeled wt) and w1118; kdm510424, #9lacZ; + (labeled kdm5−/−. * p<0.01. (G) Generation of wildtype clones in a wildtype background in larval wing imaginal disc. Genotype is hs-FLP122/+; FRT40A/FRT40A ubi-GFP. Areas circled with dashed line have two copies of ubi-GFP (twin spots). Areas shown with a solid line have no GFP. (H) Generation of kdm5K6801 mutant clones in wing imaginal disc. Genotype is hs-FLP122/+; kdm5K6801 FRT40A/FRT40A ubi-GFP. Areas shown with a dashed line have two copies of GFP (twin spots). Areas circled with sold line are homozygous mutant for kdm5K6801. (I) Quantitation of clone area in E (wt; N = 6) and F (kdm5 mutant; N = 8). ns = not significantly different. We also tested whether adult flies with reduced levels of KDM5 were sensitive to conditions of oxidative stress. To do this, we used adult flies that were transheterozygous for kdm510421 and kdm5K06801. While kdm510424 and kdm5K06801 are both homozygous lethal [22], combining these alleles decreases KDM5 protein levels 70–80% and adults eclose at 50% of the expected frequency (Figure 2B; Table S1) [13], [15]. As shown in Figure 2C and Figure S3A for females and males, respectively, kdm510424/K06801 mutant adults die significantly faster than wildtype controls when treated with paraquat. kdm5 mutant larvae and adults therefore show similar oxidative stress-sensitivity phenotypes. It should be noted that although kdm510424/K06801 adults survive to the same degree as wildtype for the first five days in non-stressed conditions, these animals are shorter-lived than wildtype (Figure S3B, C). To confirm a role for KDM5 in oxidative stress resistance, we used adult-specific RNAi-mediated knockdown, as this decreases lifespan in non-stressed conditions by only 7% compared to 87% for kdm510424/K6801 (Figure S4A, B). Because ubiquitous expression of KDM5 RNAi transgenes throughout development causes lethality, we combined Actin-Gal4 with a ubiquitously expressed temperature sensitive Gal4 inhibitor Gal80TS to control transgene expression (Gal80TS; ActTS when combined) [47]. By crossing KDM5 RNAi transgenes (or a control GFP RNAi transgene) at 18°C, transgene expression is kept off, but is activated by shifting adults to 25°C. Like kdm510242/K6801 mutants, ubiquitous expression of a KDM5 RNAi transgene during adulthood resulted in sensitivity to paraquat (Figure S4C). Control adults maintained at 18°C, at which temperature the RNAi transgene is not expressed, did not show this phenotype (Figure S4D, E). Because mutations in other genes (e.g. Foxo and Nrf2) [48], [49] that cause sensitivity to oxidative stress also affect sensitivity to other stressors, we also tested whether KDM5 knockdown adults were sensitive to treatment with a xenobiotic. Adult specific RNAi-mediated knockdown of KDM5 resulted in sensitivity to the well-characterized insecticide DDT (dichloro-diphenyl-trichloroethane) (Figure S4F). KDM5 is therefore essential for survival in response to environmental conditions of oxidative and xenobiotic stress.
Reducing KDM5 elevates levels of oxidized proteins and increases mutation frequency
Increased cellular levels of oxidative stress can cause cellular dysfunction by oxidizing proteins and DNA [50]. We therefore examined overall levels of protein oxidation in kdm5 mutant larvae and found that they have ∼2-fold increase in the levels of carbonyl groups that are introduced into proteins by oxidative reactions (Figure 2D). We also tested whether kdm5 mutant larvae have an increased nuclear DNA mutation frequency using a lacZ mutation transgene (Figure 2E) [47], [51]. Using this reporter, we found that kdm5 mutant larvae have a 2-fold higher mutation frequency than genetic background-matched controls (Figure 2F). Cells lacking KDM5 therefore experience increased oxidative stress even in the absence of any exogenous source of stress and this is toxic to both proteins and DNA. Importantly, kdm5 mutant larvae occur at approximately at the expected Mendelian ratio and kdm5 mutant cells do not show any overt growth phenotype (Figure 2G–I). The phenotypes we observe in kdm5 mutant larvae are therefore unlikely to be downstream consequences to growth and/or proliferation defects.
KDM5 physically and genetically interacts with Foxo, an established oxidative stress response transcription factor
The 16 oxidative stress genes shown in Figure 1E all have binding site(s) for Foxo, which is a transcription factor known to be integral to responding to conditions of oxidative stress [52]. One gene, 4E-BP is a well-established direct Foxo target gene [53], [54]. The remaining genes are candidate Foxo targets based on their downregulation in foxo mutant larvae microarrays or being bound by Foxo binding using ChIP-chip from wildtype larvae or ChIP-seq from adults [37], [55]–[57]. To confirm that these were Foxo regulated genes, we examined their expression in foxo21 and foxoΔ94 homozygous mutant larvae and found that 14 of the 16 genes tested were downregulated in both mutant strains (Figure 3A). Thus 13 oxidative stress sensitivity genes tested showed similar downregulation in kdm5 and foxo mutant larvae. In a similar manner to kdm5 mutant and knockdown adults, foxo mutant flies have been shown to be sensitive to conditions of oxidative stress [53] (Figure S5A). Further confirming their similarities, kdm5 and foxo mutant larvae display similar rates of death when subjected to paraquat-mediated oxidative stress (Figure 3B).
Fig. 3. KDM5 and Foxo regulate a common set of genes and genetically and physically interact. 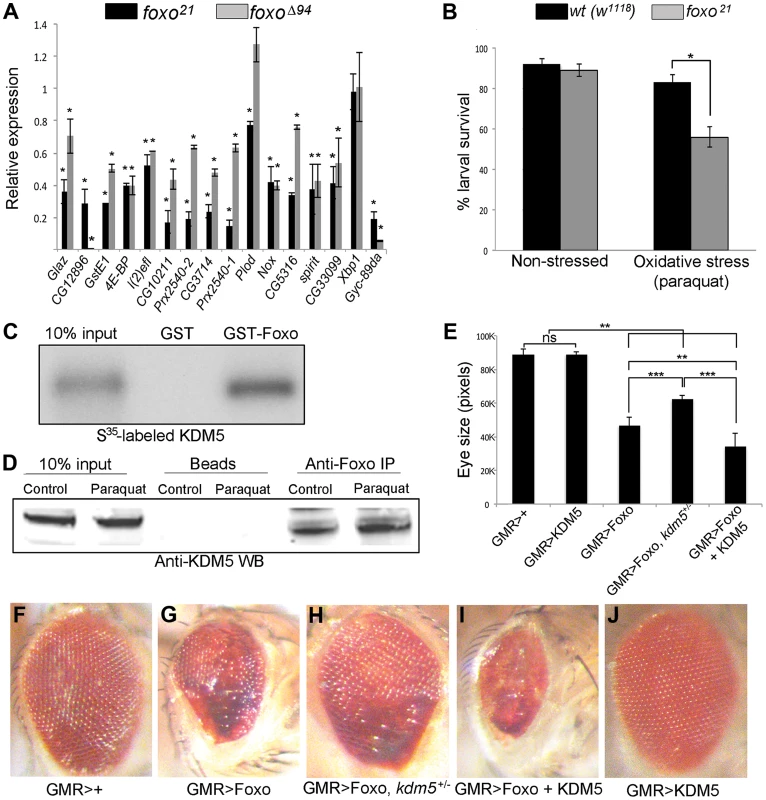
(A) Real-time PCR showing levels of gene expression in foxo21 or foxoΔ94 homozygous 3rd instar female larvae relative to wildtype controls (w1118). foxo21 was maintained as a homozygous stock while foxoΔ94 was generated by intercrossing heterozygous parents, so potentially contains some maternally derived Foxo. * p<0.05. (B) Survival of wildtype (w1118) and foxo21 female larvae in 5% sucrose (left) or with 20 mM paraquat in 5% sucrose (right) for six hours. (C) In vitro binding assay showing that S35-labeled full length KDM5 binds to GST-Foxo but not to GST alone. (D) in vivo interaction between Foxo and KDM5. Foxo protein was immunoprecipitated from S2 cell nuclear extract from cells grown in vehicle or oxidative stress conditions (20 mM paraquat for six hours). Levels of KDM5 in Foxo immunoprecipitates were then examined by Western blot and compared to beads alone. (E) Quantitation of genetic interaction between kdm5 and foxo using total eye area (>10 eyes per genotype) as measured in pixels using ImageJ. Error bars indicate standard deviation. Significance was determined using a Student's t-test. ** p<0.001, *** p<0.0001. ns = not significant. (F–J) Representative images of the genetic interaction between Foxo and KDM5 in 15 day old flies. (F) Control flies are GMR-Gal4 crossed to wildtype (w1118) and (G) GMR-Gal4 combined with UAS-Foxo showing rough eye phenotype. (H) GMR-Gal4 combined with UAS-Foxo and heterozygous for kdm5K6801 showing suppression of GMR>Foxo eye phenotype. (I) GMR-Gal4 combined with UAS-Foxo and UAS-KDM5 showing eye phenotype enhancement. (J) GMR-Gal4 crossed to UAS-KDM5. Based on the gene expression and phenotypic similarities caused by loss of KDM5 and Foxo, we tested whether these two proteins form a complex. In vitro transcribed and translated S35-labeled KDM5 bound robustly to GST-Foxo but did not bind to GST alone, demonstrating that these proteins physically interact (Figure 3C; Figure S5B). KDM5 and Foxo also interact in vivo, as immunoprecipitating Foxo from S2 cell nuclear extracts co-precipitated KDM5 in non-stressed cells and in cells subjected to increased oxidative stress through treatment with paraquat (Figure 3D; Figure S5C).
To assess the general requirement for KDM5 in Foxo function, we tested whether they genetically interact using an adult eye phenotype induced by overexpression of Foxo (Figure 3E–J). GMR-Gal4-mediated overexpression of Foxo causes an adult eye phenotype due to a reduced cell number and cell size [53], [58] (Figure 3F, G). This eye phenotype was suppressed by genetically reducing KDM5 levels using kdm5 heterozygotes (kdm5K06801 or kdm510424) (Figure 3H). Conversely, co-overexpressing Foxo and KDM5 resulted in a more severe eye phenotype than Foxo alone (Figure 3I). KDM5 overexpression alone did not result in any detectable adult phenotype, nor did kdm5 heterozygous flies (Figure 3J) [13]. These data for the first time demonstrate that KDM5 is required for Foxo function in vivo.
KDM5 primarily affects the level of oxidative stress genes in non-stressed conditions
The sensitivity of kdm5 mutants to conditions of oxidative stress may be due to their already reduced expression of oxidative stress resistance genes. KDM5 may also be required for the activation of these genes in response to acute oxidative stress conditions. For these analyses, we focused on 13 genes that were downregulated in kdm5 and foxo mutant larvae. We first tested wildtype larvae to determine which of these genes was activated in response to paraquat treatment. Eight of the 13 genes tested were induced by paraquat in wildtype larvae (Figure S5D), and we further characterized the role of the KDM5/Foxo complex in the regulation of six of these that showed the most consistent activation.
To determine the requirement for KDM5 and Foxo for stress-mediated gene activation we treated wildtype, kdm5 and foxo mutant larvae with paraquat and examined the expression of KDM5-Foxo target genes (Figure 4A). These data revealed that the primary role for KDM5 and Foxo is in maintaining the endogenous expression of oxidative stress resistance genes and not their exogenous oxidative-stress mediated activation. Three genes, 4E-BP, l(2)efl and spirit, were activated in response to paraquat but to lower maximal levels of expression in kdm5 and foxo mutants. KDM5 and Foxo are therefore required for the baseline expression of these genes, but are not essential for their activation in response to stress conditions. Conversely, one gene (CG10211) was not activated in paraquat-treated kdm5 or foxo mutants, thus this gene requires KDM5 and Foxo for its expression in both non-stressed and stressed conditions. Interestingly, two genes, the peroxiredoxin Prx2540-2 and the predicted DNA repair enzyme CG5316 were activated by paraquat in kdm5 mutant larvae but not in foxo mutants. This suggests that while KDM5 and Foxo are required for endogenous levels of expression, only Foxo is required for their paraquat-mediated activation.
Fig. 4. KDM5 directly regulates oxidative stress resistance genes. 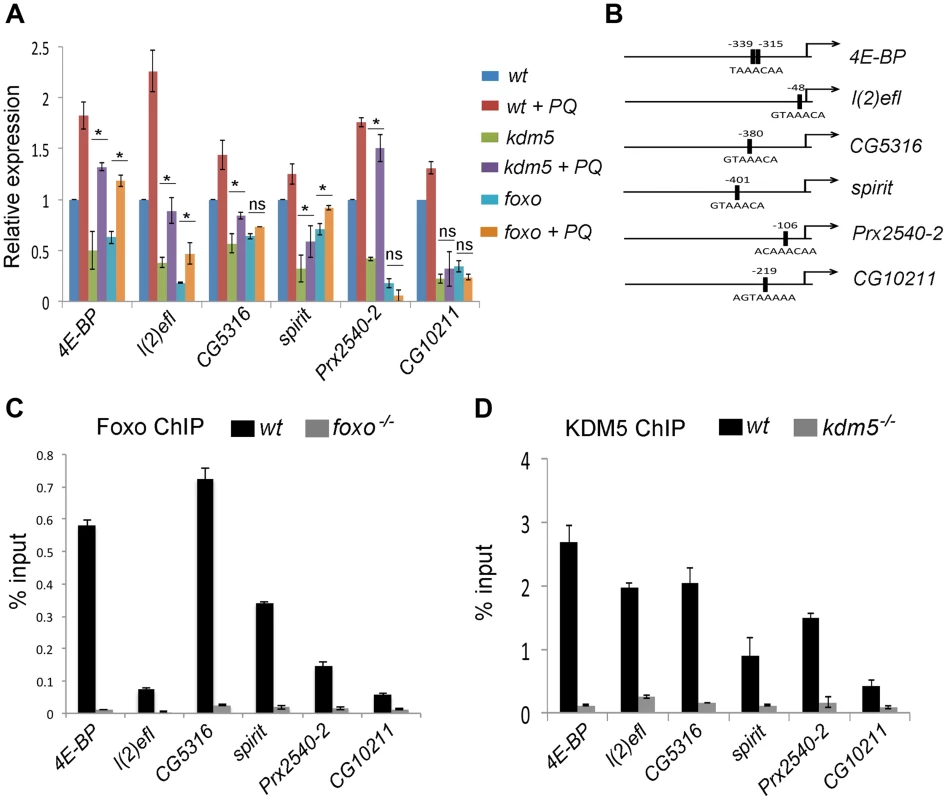
(A) Real-time PCR analyses of 4E-BP, l(2)efl, CG5316, spirit, Prx2540-2 and CG10211 from wildtype (w1118), kdm5K6801 mutant and foxo21 mutant 3rd instar larvae in 5% sucrose or 20 mM paraquat/5% sucrose for six hours (oxidative stress). (B) Schematic of the promoters of 4E-BP, l(2)efl, CG5316, spirit, Prx2540-2 and CG10211 showing the position of the Foxo binding site (FHREs; black boxes). Primers surrounding these sites were used for ChIP analyses shown in parts C and D. (C) ChIP analyses of 4E-BP, l(2)efl, CG5316, spirit, Prx2540-2 and CG10211. Anti-Foxo ChIP is shown in black bars while control anti-Foxo ChIP from foxo21 homozygous mutant larvae is shown in grey. Data are shown as % of input DNA. All six genes tested show significant attenuation of anti-Foxo ChIP signal (p<<0.01). (D) ChIP analyses of 4E-BP, l(2)efl, CG5316, spirit, Prx2540-2 and CG10211. Anti-KDM5 ChIP is shown in black bars while control anti-KDM5 ChIP from kdm5K6801 homozygous mutant larvae is shown in grey. Data are shown as % of input DNA. All six genes tested show significant attenuation of anti-KDM5 ChIP signal (p<<0.01). KDM5 and Foxo bind to the same promoter region of co-regulated genes
Based on the gene expression similarities and the genetic and physical interaction between KDM5 and Foxo, we predicted that these proteins act together to regulate gene expression. To test this, we used ChIP to confirm that Foxo bound to the predicted Forkhead response element (FHRE) region within the promoters of six oxidative stress resistance genes in larvae (Figure 4B). Foxo bound to the FHRE promoter region of all six genes using two independent anti-Foxo antibodies and using foxo mutant larvae and IgG alone as a negative controls (Figure 4C; Figure S6A, B). It should be noted that while the Foxo ChIP signal to the FHRE regions of l(2)efl and CG10211 are relatively weak (0.1% input), they are significantly attenuated in foxo mutant larvae, demonstrating that this signal is Foxo-specific. Consistent with KDM5 functioning with Foxo at these promoters, anti-KDM5 ChIP showed that KDM5 binds to the same FHRE regions as Foxo in wildtype larvae but not kdm5 mutants (Figure 4D). Importantly, Foxo and KDM5 ChIP analyses of control upstream and downstream regions of these genes revealed background levels of binding that were unaltered in foxo or kdm5 mutants, respectively (Figure S7A–C). To independently confirm KDM5 binding, we used a genomic rescue strategy to generate an epitope-tagged form of KDM5. This HA-tagged form of KDM5 is expressed at endogenous levels and rescues kdm5 mutants in a similar manner to a wildtype non-tagged version of this transgene [15] (Figure S8A, B). Like our analyses using anti-KDM5, anti-HA ChIP showed enrichment to the FHRE region of target promoters compared to an IgG specificity control (Figure S8C–G).
KDM5 is required for efficient Foxo promoter recruitment
To address the mechanism by which loss of KDM5 affects Foxo-regulated oxidative stress resistance genes, we first asked whether it was due to alterations in abundance. As shown in Figure 5A and B, levels of Foxo and KDM5 are not altered in kdm5 and foxo mutants, respectively. The changes to gene expression observed are therefore not simply due to the absence of these proteins. We next tested whether KDM5's well-established H3K4me3-directed demethylase activity played a role in the regulation of oxidative stress resistance genes. To do this, we used our previously generated fly strain that specifically lacks KDM5-dependent demethylase activity [15]. Consistent with our previous observation that global levels of H3K4me3 are increased in demethylase-inactive KDM5 mutants, the non-KDM5 target puc and the KDM5 target genes 4E-BP, l(2)efl, CG5316, spirit and CG10211 all showed increased promoter proximal H3K4me3 (Figure 5C). Prx2540-2 was the only gene examined that did not show increased levels of H3K4me3, suggesting that not all promoters are equally affected by the loss of KDM5-dependent demethylase activity. Despite increased levels of H3K4me3 that is associated with actively transcribed genes, six of seven genes were expressed at wildtype levels in KDM5 demethylase inactive larvae (Figure 5D). The one gene with increased expression (spirit) may be particularly sensitive to increased levels of H3K4me3 (Figure 5D). These data show that the demethylase activity of KDM5 is not required for the activation of KDM5-Foxo oxidative stress target genes. These expression data are consistent with our previous observation that, unlike KDM5 knockdown flies, demethylase inactive flies are not sensitive to conditions of oxidative stress [15].
Fig. 5. The H3K4me3 demethylase activity of KDM5 is not required for it to regulate KDM5-Foxo target genes. 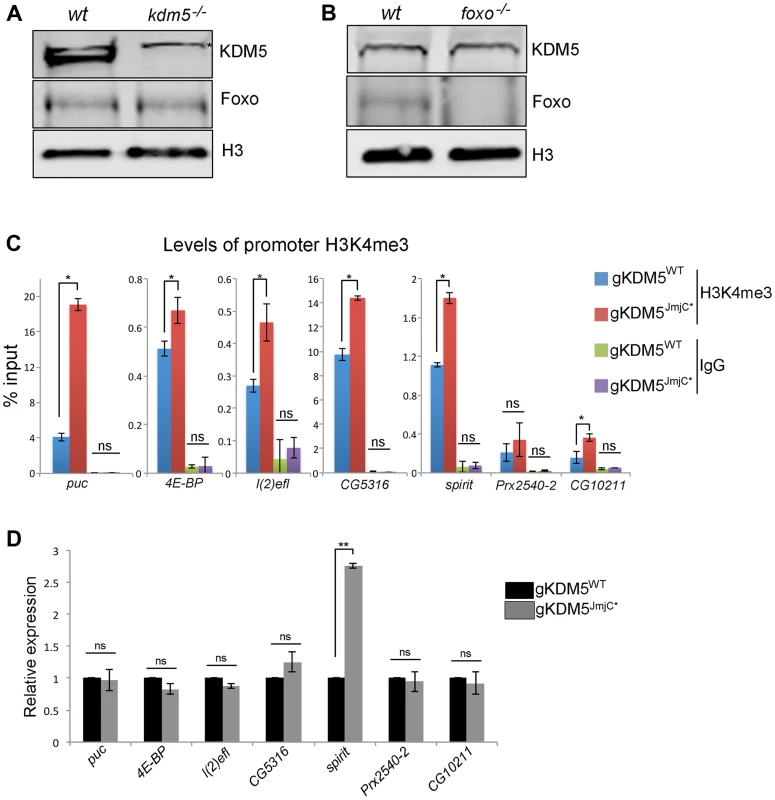
(A) Western blot showing levels of KDM5, Foxo and the loading control histone H3 in wildtype (w1118) and kdm5 mutant (kdm5K6801) 3rd instar wing imaginal discs (10 discs per lane). (B) Western blot showing levels of KDM5, Foxo and histone H3 in wildtype (w1118), and foxo21 mutant 3rd instar wing discs. (C) H3K4me3 ChIP to the FHRE region of puc, 4E-BP, l(2)efl, CG5316, spirit, Prx2540-2 and CG10211 promoters in lid10424/lid10424; gKDM5/gKDM5 (blue) and lid10424/lid10424; gKDM5JmjC*/gKDM5JmjC* (red) larvae. * p<0.05. IgG is included as a negative control. (D) Real-time PCR showing expression levels in wildtype (lid10424/lid10424; gKDM5/gKDM5 (black) and lid10424/lid10424; gKDM5JmjC*/gKDM5JmjC* (grey) 3rd instar larvae. Data are normalized to rp49 expression and changes to expression in demethylase inactive larvae are shown relative to lid10424/lid10424; gKDM5/gKDM5 control larvae. ** p<0.01. One means by which KDM5 is known to activate transcription is by inhibiting HDAC1 to increase levels of promoter histone H3 acetylation [18], [19]. To determine if KDM5-mediated HDAC1 inhibition plays a role in the activation of KDM5-Foxo target genes, we tested whether promoter proximal histone H3 acetylation was altered. As shown in Figure S6C, kdm5 mutant larvae have unaltered levels of histone H3 acetylation at four targets and slightly elevated at the other two. Because KDM5-Foxo targets did not show a consistent pattern of histone acetylation, these genes are likely to be regulated in an HDAC1-independent manner.
We next tested whether KDM5 is required for Foxo promoter recruitment. Significantly, Foxo binding to the six KDM5-Foxo co-regulated genes was attenuated in kdm5 mutant larvae using two independent anti-Foxo antibodies (Figure 6A and Figure S6B). This is not because Foxo was generally unable to bind DNA in kdm5 mutants, as its binding to two non-KDM5-regulated target promoters, InR and puc, was not affected. We also found that KDM5 promoter binding was reduced in foxo mutant larvae (Figure 6B). KDM5 and Foxo are therefore reciprocally required for recruitment to the promoters of KDM5-Foxo co-regulated genes.
Fig. 6. KDM5 is required for efficient recruitment of Foxo to a subset of its target promoters. 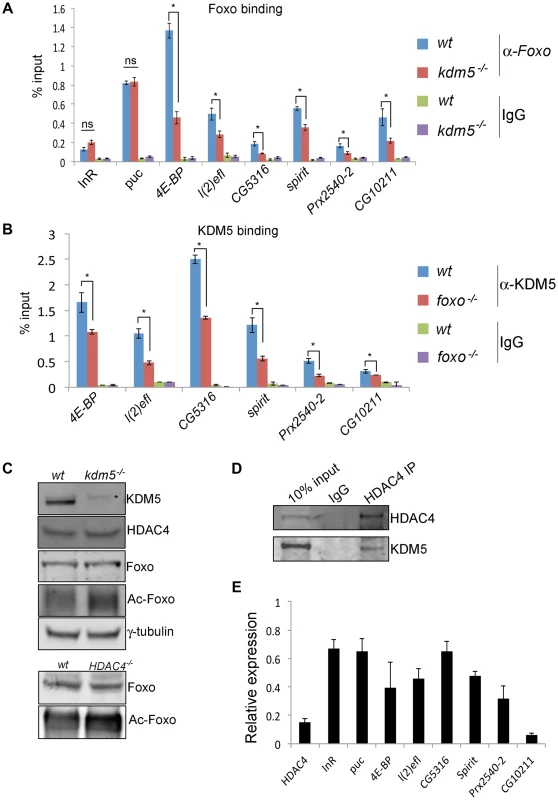
(A) ChIP using anti-Foxo [86] to the FHRE region of the InR, puc, 4E-BP, l(2)efl, CG5316, spirit, Prx2540-2 and CG10211 promoters in wildtype (w1118) and kdm510424/K6801 mutant larvae. * p<0.05. (B) KDM5 ChIP to the FHRE in the InR, puc, 4E-BP, l(2)efl, CG5316, spirit, Prx2540-2 and CG10211 promoters in wildtype (w1118) and foxo21 mutant 3rd instar larvae. * p<0.05. IgG is included as an additional control for non-specific binding in A and B. (C) (top) Levels of KDM5, HDAC4, Foxo, acetylated Foxo and the loading control γ-tubulin in wildtype and kdm5K6801 mutant larvae (anterior half of 3rd instar larvae). Levels of acetylated Foxo were determined by immunoprecipitating Foxo from larval extract and probing Western blot with an anti-acetyl-lysine antibody. * indicates non-specific band in KDM5 blot. (bottom) Levels of Foxo and acetylated Foxo in HDAC4KG09091 mutant larvae. (D) Immunoprecipitating HDAC4 from larval extracts efficiently pulls down HDAC4 and also co-precipitates KDM5. Lysate used was from 3rd instar larvae. (E) Real time PCR showing levels of HDAC4, InR, puc, 4E-BP, l(2)efl, CG5316, spirit, Prx2540-2 and CG10211 mRNA in HDAC4KG09091 homozygous 3rd instar larvae showing that all of these genes are significantly downregulated. All genes shown were significantly downregulated (p<0.05). Because Foxo DNA binding can be inhibited by acetylation [59], we tested whether KDM5 was a novel effector of this posttranslational modification. While wildtype and kdm5 mutant larvae had similar overall levels of Foxo, levels of acetylated Foxo were increased (Figure 6C, Figure S9). In both flies and mammalian cells HDAC4 can deacetylate Foxo [60], [61]. Because kdm5 mutant larvae have wildtype levels of HDAC4, the elevated levels of Foxo acetylation in these animals was not simply due to a deficiency in this enzyme (Figure 6C; Figure S9). We therefore tested whether HDAC4 interacts with KDM5, as human HDAC4 and KDM5B have been previously observed to form a complex [20]. Immunoprecipitating HDAC4 from larval extract co-precipitated KDM5, demonstrating that these proteins interact in vivo (Figure 6D; Figure S9). As previously observed [44], larvae homozygous for the HDAC4KG09091 hypomorphic mutation show increased levels of acetylated Foxo (Figure 6C). Moreover, we find that these HDAC4 mutant larvae show decreased expression of KDM5-Foxo target genes, consistent with HDAC4 functioning at these promoters with KDM5/Foxo (Figure 6E). Significantly however, we find that reducing levels of HDAC4 also decreases the expression of the Foxo targets InR and puc that are not KDM5-regulated (Figure 6E). These data suggest that KDM5 may be a component of a subset of all HDAC4-Foxo complexes that affects acetylation and promoter binding to a subset of target genes (Figure 7).
Fig. 7. Model for KDM5 function. 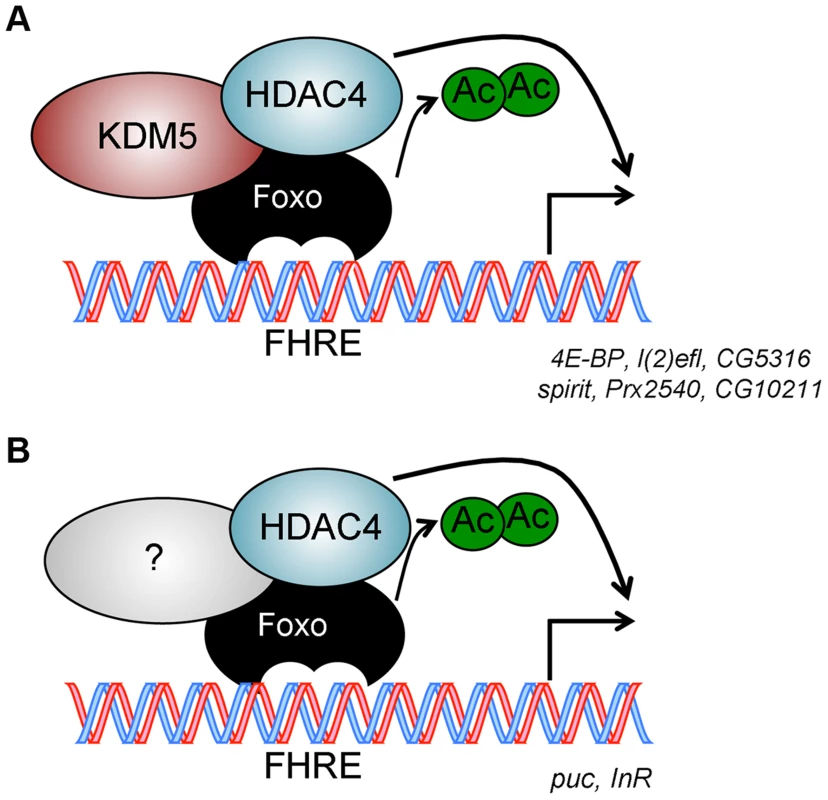
(A) KDM5-Foxo co-regulated genes are a subset of all Foxo regulated genes. We propose that at these stress response-related promoters, KDM5 facilitates the deacetylation of Foxo (and activation of its DNA binding activity) by binding to HDAC4. (B) Because HDAC4 is required at additional Foxo-regulated genes that are not affected by loss of KDM5, we propose that HDAC4-Foxo complexes that lack KDM5 regulate different targets than KDM5-HDAC4-Foxo complexes (e.g. InR and puc). Additional, as yet uncharacterized, proteins (shown as unlabeled shapes in A and B) are likely to be present in both KDM5-HDAC4-Foxo and HDAC4-Foxo complexes. See text for details. Discussion
Here we demonstrate a new function for KDM5 whereby it influences cellular levels of oxidative stress, at least in part by affecting Foxo transcription factor recruitment to a subset of its target promoters. Based on microarray data carried out on kdm5 mutant wing imaginal discs, we found that KDM5 transcriptionally regulates the expression of a number of genes implicated in regulating cellular redox state. The dysregulation of oxidative stress resistance genes in the absence of KDM5 correlates with elevated levels of oxidized proteins and increased mutation frequency. In addition, reducing levels of KDM5 in larvae or adults resulted in increased sensitivity to environmental oxidizers such as paraquat. Consistent with these data, KDM5 genetically and physically interacts with the established stress-resistance transcription factor Foxo, and these proteins co-occupy FHRE response elements within promoters. In the absence of KDM5, Foxo recruitment to a subset of its target genes is attenuated. Significantly, this correlates with higher levels of Foxo acetylation, which is known to affect Foxo DNA binding. This leads us to propose a model in which KDM5 acts to facilitate Foxo DNA binding by recruiting HDAC4, resulting in transcriptional activation of a subset of stress resistance genes.
Transcriptional regulation of redox states by KDM5
A total of 901 genes were up - or downregulated more than 1.5-fold in our microarray analyses of kdm510424 mutant wing imaginal discs. All genes tested as part of our validation were similarly affected in kdm510424 mutants in addition to kdm510424/kdm5K6801 larvae, suggesting that our expression data are robust. Interestingly, our data are significantly different to microarray analyses using wing discs from KDM5 knockdown animals recently reported, in which only 42 genes were altered more than 1.5-fold (11 upregulated and 31 downregulated) [62]. Indeed, none of the 42 genes that were affected in these KDM5 knockdown animals were altered in our analyses of kdm5 mutant wing discs. These differences could be related the fact that we used four-fold more RNA for our transcriptome analyses, allowing us to better detect moderate changes to gene expression. Another contributing factor could be differences due to using KDM5 knockdown animals rather than genetic mutants. Whatever the basis for the disparity, our microarrays enabled us to link KDM5 to the regulation of cellular redox state for the first time.
Our observation that KDM5 affects cellular levels of oxidative stress is particularly relevant in light of evidence that dysregulated redox state contributes to the pathogenesis of several human diseases [5], [63]. Human KDM5A and KDM5B are overexpressed in many forms of cancer [3], [64]. While the role of KDM5A remains less characterized, KDM5B is specifically implicated in the survival of cancer “stem cells” [23], [24]. By promoting the survival and slow proliferation of this small subset of cells, KDM5B overexpression creates tumors that are difficult to effectively treat with standard therapies that target rapidly dividing cells. Because transformed cells have elevated levels of reactive oxygen species (reviewed in [63]), increasing the levels of KDM5A and/or KDM5B may promote cell survival. It is interesting to note that targeting KDM5-dependent demethylase activity has been proposed as a new therapeutic strategy for treating KDM5A/B-overexpressing tumors [3]. However, KDM5A was recently shown to promote breast cancer progression and metastasis in a demethylase-independent manner [65]. Taken with our data demonstrating that KDM5 acts to regulate cellular oxidative stress independently of its enzymatic activity, we suggest that approach may not yield any significant clinical improvement for these patients.
To-date, 22 mutations in the human KDM5C paralog have been found in patients with X-linked intellectual disability [29]–[32]. Increased oxidative stress has been proposed to contribute to the cognitive phenotypes of diseases including Down syndrome [34], Rett syndrome [66] and Alzheimer's disease [67]–[69]. In light of our analyses, it is possible that oxidative stress-mediated cellular damage may contribute to the intellectual impairment and other phenotypes associated with by loss of KDM5C in humans such as seizures. In the absence of KDM5C, increased oxidative damage to proteins and DNA may ultimately result in cellular dysfunction and/or death. Because KDM5C is most highly expressed in neuronal cells, other KDM5 family proteins may be unable to compensate for its loss, resulting in this cell type being vulnerable to the loss of this paralog [70]. Based on this model, we are currently examining the role of KDM5-mediated oxidative stress resistance and its relationship to learning and memory defects.
KDM5-mediated regulation of Foxo promoter recruitment
Our data show that KDM5 regulates levels of cellular oxidative stress, at least in part through its interaction with Foxo. Indeed, levels of Foxo family proteins in human cells may therefore influence the tumorigenic and cognitive phenotypes observed as a result of KDM5 protein dysfunction. Importantly, KDM5 does not affect Foxo recruitment to all of its known target genes. This is likely to be because KDM5 is present in a subset of Foxo complexes, so only alters the acetylation of a specific pool of Foxo proteins. Consistent with this model, we find that two genes that require HDAC4 and Foxo for their activation, InR and puc, were not downregulated in kdm5 mutants. Thus HDAC4 likely plays a more global role in Foxo acetylation than KDM5. While it remains to be determined why some Foxo targets and not others require a KDM5, the observation that Foxo requires different co-factors at distinct targets is not unprecedented. For example, Foxo requires the SWI/SNF chromatin remodeling complex at about a third of its target genes [71]. Different chromatin contexts and nuclear microenvironments may therefore require Foxo to utilize different co-factors. Promoter-specific requirements for co-factors have also been described for other transcription factors (e.g. Myc and Hsf) [72], [73]. In addition to facilitating Foxo deacetylation and activation of DNA binding capabilities, KDM5's may play additional roles in the activation of KDM5-Foxo targets. Specifically, KDM5 may use the ability to recognize specific chromatin contexts to facilitate transcriptional activation; the PHD1 of KDM5 recognizes histone H3 that is unmethylated at lysine 4 (H3K4me0) and PHD3 recognizes histone H3 that is di - and trimethylated at lysine 4 (H3K4me2/3) [15], [64].
Based on our observation that KDM5 interacts with HDAC4, we suggest that it by recruiting this enzyme that KDM5 affects Foxo acetylation (Figure 7). Alternatively, KDM5 could affect the deacetylase activity of HDAC4 within the KDM5-Foxo complexes to regulate levels of Foxo acetylation, since KDM5 can inhibit HDAC1 [18], [19]. Even though KDM5 and HDAC4 interact, it is also possible that KDM5 affects Foxo acetylation through an HDAC4-independent mechanism. For example, Sir2 has also been shown to deacetylate Foxo [74], [75], and the relative functions of HDAC4 and Sir2 remain unclear. KDM5 could therefore influence Foxo acetylation through Sir2. Foxo acetylation is also positively regulated by the lysine acetyltransferase CBP/p300, thus it is also formally possible that KDM5 acts by negatively regulating CBP/p300 activity.
KDM5-dependent and independent Foxo complexes
The KDM5-Foxo complex is also likely to be important for cellular processes in addition to oxidative stress resistance. For example, based on the genetic interaction we observed between kdm5 and foxo in the eye, KDM5 and Foxo may act together to regulate cell size and number in some circumstances. In addition, the KDM5-Foxo complex may also function to influence aging. Levels of Foxo influence lifespan in a number of species: reducing Foxo shortens lifespan while increasing Foxo extends it [76]. In a similar manner to foxo mutant flies, the hypomorphic kdm5 allele combination of kdm510424/kdm5k6801 dramatically shortens, and adult specific KDM5 knockdown slightly shortens, lifespan. This is likely to be a conserved function of KDM5 family proteins, as loss of KDM5 (Rbr-2) in c. elegans shortens lifespan and its overexpression extends lifespan [77], [78]. It will therefore be important to determine whether adult-specific overexpression of KDM5 extends lifespan in Drosophila, and whether this is dependent on levels of Foxo. Because there is a correlation between redox state and aging [79], effects of the KDM5/Foxo complex on lifespan and aging could be mediated through their regulation of oxidative stress resistance genes.
Although kdm5 and foxo mutants phenocopy each other in some regards, both KDM5 and Foxo also have roles outside this complex. This is based on the observations that not all Foxo-regulated oxidative stress resistance genes are affected by loss of KDM5 and that while kdm5 is an essential gene, foxo null mutants are homozygous viable [22], [80]. Based on our finding that KDM5 affects Foxo acetylation and promoter recruitment, it is tempting to speculate that one of the reasons that KDM5 is essential for viability is that it acts through a similar mechanism to influence the activity of other transcription factors. Many transcription factors are acetylated, including Myc, which we have shown interacts with KDM5, in addition to p53, GATA and STAT family proteins [59], [81]–[83]. KDM5 could therefore interact with these factors to impact a broad range of essential cellular functions.
Materials and Methods
Drosophila stocks
All fly stocks were maintained at 25°C on standard medium, 60% humidity, and a 12 hour light/dark cycle. kdm5 mutant alleles kdm510424, kdm5K06801, UAS-KDM5RNAi (TRIP line 35706), UAS-Foxo, Actin-Gal4, tubulin-Gal80ts, FRT40A, hs-FLP122, GMR-Gal4, foxoΔ94 (maintained as a heterozygous stock over TM6B) and HDAC4KG09091 are publically available and were obtained from the Bloomington stock center. foxo21 was obtained from Marc Tatar [84] and was maintained as a homozygous stock. UAS-KDM5RNAi was generated by cloning an inverted repeat of KDM5 into the BamHI site of pMF3 using the primers gcggatccgtacatgcagcgtcagcggcaac (BamHI site underlined) and gcgaattccgcattattgcctccagtagctg (EcoRI site used to join inverted repeats underlined). Control UAS-GFPRNAi transgenic flies were generated by cloning an inverted repeat as a BamHI fragment using the primers gcggatccctggaaaactacctgttccatg (BamHI site underlined) and gcgaattcgttcatccatgccatgtgtaatc (EcoRI site that joins two repeats underlined).
To generate kdm5 mutant clones, we crossed hs-FLP122; FRT40A ubi-GFP virgins to +; kdm5K6801 FRT40/CyO males. Progeny were heat shocked for 45 min at 48 hours AED, and larvae were dissected at wandering 3rd instar stage. As a control, hs-FLP122; FRT40A ubi-GFP virgins were crossed to FRT40/CyO males. Clone size was determined by quantifying the total area of dark GFP positive (twin spot): GFP negative (control wt or kdm5 mutant cells) using Image J.
The HA-tagged KDM5 genomic rescue transgene was generated by inserting the coding sequence for three HA tags (YPYDVPDYA) at the 3′end of the kdm5 open reading frame after removal of the endogenous stop codon. This kdm5-HA open reading frame fragment was then cloned downstream of the kdm5 promoter in the pCasper 4 vector as described for our non-tagged genomic rescue transgene [15]. kdm5-HA transgenic flies were crossed into a kdm5 (kdm510424 or kdm5K06801) mutant background. Insertions on the 3rd chromosome and X chromosome were used and behaved indistinguishably. Transgenic flies were generated by “thebestgene.com”.
Antibodies and Westerns
The KDM5 antibody has been described previously [13], [85], anti-Foxo antibodies were obtained from Marc Tatar (Brown University; used for ) and from Cosmo Bio USA (used for Figure 6) [86], anti-H3K4me3 and anti-histone H3 were from Active Motif, γ-tubulin from Cell Signaling, anti-pan acetylated H3 from Upstate and anti-HA from Invitrogen. To detect Drosophila HDAC4, we used an antibody raised to human HDAC4, 5 and 9 from Novus. In Drosophila, our Western analyses show that this antibody recognizes a single band of the predicted molecular weight for HDAC4 (125 kDa) and that this co-migrates with a transfected FLAG-tagged form of HDAC4 (Figure S10). We therefore conclude that this antibody is specific to HDAC4 in flies. Western analysis was carried out using PVDF and standard protocols, infrared conjugated secondary antibodies (LiCOR) and Odyssey scanner and software. Signal intensities were quantitated using LiCOR software.
In vitro and in vivo binding assays
In vitro GST-fusion protein binding assays were performed as described previously [87]. For immunoprecipitations of Foxo, S2 cell nuclei with or without 20 mM paraquat treatment were isolated and fractionated using the Nuclear Extract Kit (Active Motif). Nuclear extract was dilute to 1 ml in cold immunoprecipitation buffer (20 mM HEPES, PH 7.5, 50 mM KCl, 0.05% Triton X-100, 2.5 mM EDTA, 5 mM DTT, 5% glycerol, 10 mg/ml protease inhibitor cocktail (Fisher). Extracts were pre-cleared for 30 minutes with 30 µl Protein G Sepharose (Invitrogen) in a total volume of 500 µl. After centrifugation, the supernatant was used to carry out immunoprecipitation by incubation with anti-Foxo or anti-KDM5. For immunopreciptations of HDAC4, total cell lysates were used and processed as described above for Foxo.
RT-PCR
Whole larva or dissected tissue were added to TRIZOL (Invitrogen) to extract RNA followed by cleanup using DNA-free (Ambion). 500 ng of total RNA was reverse transcribed at 42°C for 30 minutes using Verso cDNA kit (Thermo Scientific) with oligo (dt) primer to generate cDNA. Quantitative real-time PCR (qRT-PCR) analysis was performed using SYBR Green Master Mix (Thermo scientific). qRT-PCR reactions were performed in triplicate in total volumes of 10 µl containing Fast SYBR Green Master Mix, 0.25 µl of each gene-specific primer, 0.5 µl of first strand cDNA template, and nuclease free water. All qRT-PCR reactions were performed using Applied Biosystems StepOnePlus real time PCR system with the following conditions: 95°C for 15 min followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s and extension at 72 for 30 s. Data analysis was performed using StepOne software (Applied Biosystems). Fold change was calculated using s-DDCT method [88]. At least three biological replicates were used for each primer set. Primer sequences are provided in Table S2. Levels of gene expression in each sample were normalized to the expression of the housekeeping gene rp49.
Paraquat and DDT treatment
Larvae were treated with 20 mM paraquat for six hours in 5% sucrose (or sucrose alone as a control). Adults were treated with 20 mM paraquat by placing them in a vial with a paraquat-soaked (or sucrose alone) piece of whatman paper. Adults were treated with the insecticide DDT according to a previously published protocol [89].
Gene array processing and statistical analysis
Total RNA was prepared from three separate samples of 60 dissected w1118 and kdm510424 wing imaginal discs (96 hr AED) using Trizol, followed by on-column digestion of DNA using the RNeasy Mini Kit (Qiagen). RNA quantity and quality were assessed with a Beckman Coulter DU 640 spectrophotometer and Agilent 2100 Bioanalyzer, following the manufacturer's protocols. Biotinylated, single-stranded cDNA was prepared from 100 ng total intact RNA. cDNA was hybridized to Drosophila Genome 2.0 GeneChips (Affymetrix). After hybridization, GeneChips were scanned using a GeneChip Scanner 3000 (Affymetrix). CEL files were generated from DAT files using GCOS software (Affymetrix). Data were processed with Expression Console (Affymetrix) using the PLIER algorithm and Array Assist Lite software was used to generate GC-RMA files (log2 transformed) for each chip. All procedures were performed in three biological replicates at the Genome Center of the Albert Einstein College of Medicine. Fold change was calculated for kdm510424 relative to wildtype (wt; w1118). Statistical significance (p value) was calculated by Student's t-test, based on the results of three arrays from wt and kdm510424. Genes that changed by less than 1.5-fold and had a p value more than 0.05 were removed from subsequent analysis. The normalized RMA values from these genes (affected ≥1.5 fold, p<0.05) were used to perform a hierarchical cluster analysis and to construct a heat map using the Gene Cluster 3.0 and tree view software [90]. Cluster analysis groups together genes with comparable patterns of expression by employing euclidean distance metric and the Centroid linkage method. The false discovery rate (Q value) was calculated for each p-value using R [91], [92]. The accession number for microarray data is GSZ53881.
Functional interpretation of microarray data
Degree of enrichment for cellular component, biological processes and molecular functions was assessed using the Gene ontology (GO) program DAVID and Easy GO [93]–[95]. Hierarchical cluster analysis was to construct a heat map using Gene Cluster 3.0 and Java tree view software [90].
lacZ mutation analyses and protein oxidation assay
To determine lacZ mutation frequency in female larvae, #9lacZ or kdm510424, #9lacZ homozygous larvae were selected. Both the #9lacZ transgene and the kdm510424, #9lacZ recombinant chromosome were crossed into a w1118 genetic background. The lacZ transgenes, procedures for DNA extraction and mutation frequency determination have been previously described [51]. Total levels of oxidized protein was determined using “oxyblot” from Millipore according to manufacturer specifications.
Chromatin immunoprecipitation
ChIP was performed using the anterior half of 3rd instar larvae essentially as previously described [96]. Cross-linking was performed for 25 min using 1.8% formaldehyde during tissue homogenization. Chromatin extracts were sonicated using a Bioruptor (Diagenode) for 25 min (settings 30 sec on, 30 sec off, high power) to give rise to sheared chromatin with an average length of 200 to 800 bp. Immunoprecipitations were performed using 2–4 µg of anti-Foxo, anti-KDM5, anti-H3K4me3, anti-H3Ac or anti-HA. For ChIP-qPCR, triplicates from two independent biological replicates were analyzed following the ΔCt method. Data are expressed as the percentage of input chromatin precipitated for each region examined. Table S2 lists the sequences of primers used in these experiments.
Statistical analyses
The statistical significance of gene expression and ChIP binding were determined using a student's t-test (Microsoft excel). Statistical significance of lifespan data were determined with Log-rank (Mantel-cox) test using Prism (GraphPad software). Eye size of GMR>Foxo and other genotypes was determined using ImageJ and statistical significance determined using a student's t-test. R2 correlation coefficients for microarray biological replicates were generated using ArrayStar Scatter Plot (DNAStar) and are shown in Figure S1.
Supporting Information
Zdroje
1. RothbartSB, StrahlBD (2014) Interpreting the language of histone and DNA modifications. Biochim Biophys Acta 1839 : 627–43 doi: 10.1016/j.bbagrm.2014.03.001
2. MargueronR, ReinbergD (2010) Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 11 : 285–296.
3. BlairLP, CaoJ, ZouMR, SayeghJ, YanQ (2011) Epigenetic Regulation by Lysine Demethylase 5 (KDM5) Enzymes in Cancer. Cancers (Basel) 3 : 1383–1404.
4. SecombeJ, EisenmanRN (2007) The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases - The Myc connection. Cell Cycle 6 : 1324–1328.
5. Lopez-BigasN, KisielTA, DeWaalDC, HolmesKB, VolkertTL, et al. (2008) Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Molecular Cell 31 : 520–530.
6. ChristensenJ, AggerK, CloosPA, PasiniD, RoseS, et al. (2007) RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone H3. Cell 128 : 1063–1076.
7. KloseRJ, YanQ, TothovaZ, YamaneK, Erdjument-BromageH, et al. (2007) The Retinoblastoma binding protein RBP2 is a H3K4 demethylase. Cell 128 : 889–900.
8. IwaseS, LanF, BaylissP, de la Torre-UbietaL, HuarteM, et al. (2007) The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128 : 1077–1088.
9. YamaneK, TateishiK, KloseRJ, FangJ, FabrizioLA, et al. (2007) PLU-1 is a H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Molecular Cell 25 : 801–812.
10. LeeMG, NormanJ, ShilatifardA, ShiekhattarR (2007) Physical and functional association of a trimethyl H3K4 demethylase and RING6a/MBLR, a Polycomb-like protein. Cell 128 : 877–887.
11. EissenbergJC, LeeMG, SchneiderJ, IlvarsonnA, ShiekhattarR, et al. (2007) The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nature Structural & Molecular Biology 14 : 344–346.
12. LeeN, ZhangJY, KloseRJ, Erdjument-BromageH, TempstP, et al. (2007) The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nature Structural & Molecular Biology 14 : 341–343.
13. SecombeJ, LiL, CarlosLS, EisenmanRN (2007) The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes & Development 21 : 537–551.
14. KloseRJ, ZhangY (2007) Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8 : 307–318.
15. LiL, GreerC, EisenmanRN, SecombeJ (2010) Essential functions of the histone demethylase lid. PLoS Genet 6: e1001221.
16. CatchpoleS, Spencer-DeneB, HallD, SantangeloS, RosewellI, et al. (2011) PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int J Oncol 38 : 1267–1277.
17. AlbertM, SchmitzSU, KooistraSM, MalatestaM, Morales TorresC, et al. (2013) The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet 9: e1003461.
18. DiTacchioL, LeHD, VollmersC, HatoriM, WitcherM, et al. (2011) Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science 333 : 1881–1885.
19. LeeN, Erdjument-BromageH, TempstP, JonesRS, ZhangY (2009) The H3K4 Demethylase Lid Associates with and Inhibits the Histone Deacetylase Rpd3. Molecular and Cellular Biology 29 : 1401–1410.
20. BarrettA, SantangeloS, TanK, CatchpoleS, RobertsK, et al. (2007) Breast cancer associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. International Journal of Cancer 121 : 265–275.
21. KleinBJ, PiaoL, XiY, Rincon-AranoH, RothbartSB, et al. (2014) The Histone-H3K4-Specific Demethylase KDM5B Binds to Its Substrate and Product through Distinct PHD Fingers. Cell Rep 6 : 325–35 doi: 10.1016/j.celrep.2013.12.021
22. GildeaJJ, LopezR, ShearnA (2000) A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics 156 : 645–663.
23. RoeschA, Fukunaga-KalabisM, SchmidtEC, ZabierowskiSE, BraffordPA, et al. (2010) A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 141 : 583–594.
24. RoeschA, VulturA, BogeskiI, WangH, ZimmermannKM, et al. (2013) Overcoming Intrinsic Multidrug Resistance in Melanoma by Blocking the Mitochondrial Respiratory Chain of Slow-Cycling JARID1B(high) Cells. Cancer Cell 23 : 811–825.
25. OutchkourovNS, MuinoJM, KaufmannK, van IjckenWF, Groot KoerkampMJ, et al. (2013) Balancing of histone H3K4 methylation states by the Kdm5c/SMCX histone demethylase modulates promoter and enhancer function. Cell Rep 3 : 1071–1079.
26. BenevolenskayaEV (2007) Retinoblastoma binding protein 2 (RBP2) and differentiation. Biochemistry and Cell Biology-Biochimie Et Biologie Cellulaire 85 : 523–523.
27. BenevolenskayaEV, MurrayHL, BrantonP, YoungRA, KaelinWG (2005) Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Molecular Cell 18 : 623–635.
28. LinW, CaoJ, LiuJ, BeshiriML, FujiwaraY, et al. (2011) Loss of the retinoblastoma binding protein 2 (RBP2) histone demethylase suppresses tumorigenesis in mice lacking Rb1 or Men1. Proc Natl Acad Sci U S A 108 : 13379–13386.
29. JensenLR, AmendeM, GurokU, MoserB, GimmelV, et al. (2005) Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. American Journal of Human Genetics 76 : 227–236.
30. RujirabanjerdS, NelsonJ, TarpeyPS, HackettA, EdkinsS, et al. (2010) Identification and characterization of two novel JARID1C mutations: suggestion of an emerging genotype-phenotype correlation. Eur J Hum Genet 18 : 330–335.
31. SantosC, Rodriguez-RevengaL, MadrigalI, BadenasC, PinedaM, et al. (2006) A novel mutation in JARID1C gene associated with mental retardation. Eur J Hum Genet 14 : 583–586.
32. Santos-ReboucasCB, Fintelman-RodriguesN, JensenLR, KussAW, RibeiroMG, et al. (2011) A novel nonsense mutation in KDM5C/JARID1C gene causing intellectual disability, short stature and speech delay. Neurosci Lett 498 : 67–71.
33. MartinI, GrotewielMS (2006) Oxidative damage and age-related functional declines. Mech Ageing Dev 127 : 411–423.
34. PaganoG, CastelloG (2012) Oxidative stress and mitochondrial dysfunction in Down syndrome. Adv Exp Med Biol 724 : 291–299.
35. VictorinoVJ, PizzattiL, MichellettiP, PanisC (2014) Oxidative Stress, Redox Signaling and Cancer Chemoresistance: Putting Together the Pieces of the Puzzle. Curr Med Chem 21 : 3211–26.
36. ZidBM, RogersAN, KatewaSD, VargasMA, KolipinskiMC, et al. (2009) 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 139 : 149–160.
37. TelemanAA, ChenYW, CohenSM (2005) 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev 19 : 1844–1848.
38. TettweilerG, MironM, JenkinsM, SonenbergN, LaskoPF (2005) Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Genes Dev 19 : 1840–1843.
39. WoodZA, SchroderE, Robin HarrisJ, PooleLB (2003) Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 28 : 32–40.
40. RadyukSN, MichalakK, KlichkoVI, BenesJ, RebrinI, et al. (2009) Peroxiredoxin 5 confers protection against oxidative stress and apoptosis and also promotes longevity in Drosophila. Biochem J 419 : 437–445.
41. TulsawaniR, KellyLS, FatmaN, ChhunchhaB, KuboE, et al. (2010) Neuroprotective effect of peroxiredoxin 6 against hypoxia-induced retinal ganglion cell damage. BMC Neurosci 11 : 125.
42. MassudiH, GrantR, GuilleminGJ, BraidyN (2012) NAD+ metabolism and oxidative stress: the golden nucleotide on a crown of thorns. Redox Rep 17 : 28–46.
43. WalkerDW, MuffatJ, RundelC, BenzerS (2006) Overexpression of a Drosophila homolog of apolipoprotein D leads to increased stress resistance and extended lifespan. Current Biology 16 : 674–679.
44. WangMC, BohmannD, JasperH (2005) JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 121 : 115–125.
45. LinYJ, SeroudeL, BenzerS (1998) Extended life-span and stress resistance in the Drosophila mutant methuselah. Science 282 : 943–946.
46. SanchezD, Lopez-AriasB, TorrojaL, CanalI, WangX, et al. (2006) Loss of glial lazarillo, a homolog of apolipoprotein D, reduces lifespan and stress resistance in Drosophila. Current Biology 16 : 680–686.
47. GreerC, LeeM, WesterhofM, MilhollandB, SpokonyR, et al. (2013) Myc-dependent genome instability and lifespan in Drosophila. PLoS One 8: e74641.
48. SlackC, GiannakouME, FoleyA, GossM, PartridgeL (2011) dFOXO-independent effects of reduced insulin-like signaling in Drosophila. Aging Cell 10 : 735–748.
49. DengH (2014) Multiple roles of Nrf2-Keap1 signaling: regulation of development and xenobiotic response using distinct mechanisms. Fly (Austin) 8 : 7–12.
50. HwangAB, JeongDE, LeeSJ (2012) Mitochondria and organismal longevity. Curr Genomics 13 : 519–532.
51. GarciaAM, DerventziA, BusuttilR, CalderRB, PerezEJr, et al. (2007) A model system for analyzing somatic mutations in Drosophila melanogaster. Nat Methods 4 : 401–403.
52. CalnanDR, BrunetA (2008) The FoxO code. Oncogene 27 : 2276–2288.
53. JüngerMA, RintelenF, StockerH, WassermanJD, VeghM, et al. (2003) The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol 2 : 20.
54. PuigO, MarrMT, RuhfML, TjianR (2003) Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev 17 : 2006–2020.
55. AlicN, AndrewsTD, GiannakouME, PapatheodorouI, SlackC, et al. (2011) Genome-wide dFOXO targets and topology of the transcriptomic response to stress and insulin signalling. Mol Syst Biol 7 : 502.
56. BaiH, KangP, HernandezAM, TatarM (2013) Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila. PLoS Genet 9: e1003941.
57. TelemanAA, HietakangasV, SayadianAC, CohenSM (2008) Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab 7 : 21–32.
58. KramerJM, DavidgeJT, LockyerJM, StaveleyBE (2003) Expression of Drosophila FOXO regulates growth and can phenocopy starvation. BMC Dev Biol 3 : 5.
59. DaitokuH, SakamakiJ, FukamizuA (2011) Regulation of FoxO transcription factors by acetylation and protein-protein interactions. Biochim Biophys Acta 1813 : 1954–1960.
60. WangB, MoyaN, NiessenS, HooverH, MihaylovaMM, et al. (2011) A hormone-dependent module regulating energy balance. Cell 145 : 596–606.
61. MihaylovaMM, VasquezDS, RavnskjaerK, DenechaudPD, YuRT, et al. (2011) Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145 : 607–621.
62. Lloret-LlinaresM, Perez-LluchS, RossellD, MoranT, Ponsa-CobasJ, et al. (2012) dKDM5/LID regulates H3K4me3 dynamics at the transcription-start site (TSS) of actively transcribed developmental genes. Nucleic Acids Research 40 : 9493–505 doi: 10.1093/nar/gks773
63. LiX, FangP, MaiJ, ChoiET, WangH, et al. (2013) Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J Hematol Oncol 6 : 19.
64. WangGG, SongJ, WangZ, DormannHL, CasadioF, et al. (2009) Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459 : 847–851.
65. CaoJ, LiuZ, CheungWK, ZhaoM, ChenSY, et al. (2014) Histone demethylase RBP2 is critical for breast cancer progression and metastasis. Cell Rep 6 : 868–877.
66. De FeliceC, SignoriniC, LeonciniS, PecorelliA, DurandT, et al. (2012) The role of oxidative stress in Rett syndrome: an overview. Ann N Y Acad Sci 1259 : 121–135.
67. YanMH, WangX, ZhuX (2013) Mitochondrial defects and oxidative stress in Alzheimer disease and Parkinson disease. Free Radic Biol Med 62 : 90–101.
68. HuZ, ChenL, ZhangJ, LiT, TangJ, et al. (2007) Structure, function, property, and role in neurologic diseases and other diseases of the sHsp22. J Neurosci Res 85 : 2071–2079.
69. PattenDA, GermainM, KellyMA, SlackRS (2010) Reactive oxygen species: stuck in the middle of neurodegeneration. J Alzheimers Dis 20 Suppl 2: S357–367.
70. XuJ, DengX, DistecheCM (2008) Sex-Specific Expression of the X-Linked Histone Demethylase Gene Jarid1c in Brain. Plos One 3: e2553.
71. RiedelCG, DowenRH, LourencoGF, KirienkoNV, HeimbucherT, et al. (2013) DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat Cell Biol 15 : 491–501.
72. EberhardySR, FarnhamPJ (2002) Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. Journal of Biological Chemistry 277 : 40156–40162.
73. EberhardySR, FarnhamPJ (2002) Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. Faseb Journal 16: A1–a1.
74. BrunetA, SweeneyLB, SturgillJF, ChuaKF, GreerPL, et al. (2004) Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303 : 2011–2015.
75. DaitokuH, HattaM, MatsuzakiH, ArataniS, OhshimaT, et al. (2004) Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A 101 : 10042–10047.
76. WatrobaM, MaslinskaD, MaslinskiS (2012) Current overview of functions of FoxO proteins, with special regards to cellular homeostasis, cell response to stress, as well as inflammation and aging. Adv Med Sci 57 : 183–195.
77. GreerEL, MauresTJ, HauswirthAG, GreenEM, LeemanDS, et al. (2010) Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466 : 383–387.
78. AlvaresSM, MayberryGA, JoynerEY, LakowskiB, AhmedS (2013) H3K4 demethylase activities repress proliferative and postmitotic aging. Aging Cell 13 : 245–53 doi: 10.1111/acel.12166
79. OrrWC, RadyukSN, SohalRS (2013) Involvement of redox state in the aging of Drosophila melanogaster. Antioxid Redox Signal 19 : 788–803.
80. PuigO, TijanR (2005) Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes & Development 19 : 2435–2446.
81. GuW, RoederRG (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90 : 595–606.
82. GuW, ShiXL, RoederRG (1997) Synergistic activation of transcription by CBP and p53. Nature 387 : 819–823.
83. BoyesJ, ByfieldP, NakataniY, OgryzkoV (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396 : 594–598.
84. MinKJ, YamamotoR, BuchS, PankratzM, TatarM (2008) Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell 7 : 199–206.
85. ZaffranS, ChartierA, GallantP, AstierM, ArquierN, et al. (1998) A Drosophila RNA helicase gene, pitchoune, is required for cell growth and proliferation and is a potential target of d-Myc. Development 125 : 3571–3584.
86. KanaoT, VenderovaK, ParkDS, UntermanT, LuB, et al. (2010) Activation of FoxO by LRRK2 induces expression of proapoptotic proteins and alters survival of postmitotic dopaminergic neuron in Drosophila. Hum Mol Genet 19 : 3747–3758.
87. HurlinPJ, FoleyKP, AyerDE, EisenmanRN, HanahanD, et al. (1995) Regulation of Myc and Mad during epidermal differentiation and HPV-associated tumorigenesis. Oncogene 11 : 2487–2501.
88. LivakKJ, SchmittgenTD (2001) Analysis of relative gene expression data using Real-Time quantitative PCR and the 2−DDC method. Methods 25 : 402–408.
89. GronkeS, ClarkeDF, BroughtonS, AndrewsTD, PartridgeL (2010) Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet 6: e1000857.
90. de HoonMJ, ImotoS, NolanJ, MiyanoS (2004) Open source clustering software. Bioinformatics 20 : 1453–1454.
91. StoreyJD, TibshiraniR (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100 : 9440–9445.
92. StoreyJD, TibshiraniR (2003) Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol Biol 224 : 149–157.
93. Huang daW, ShermanBT, ZhengX, YangJ, ImamichiT, et al. (2009) Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics Chapter 13: Unit 13 11.
94. Huang daW, ShermanBT, LempickiRA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4 : 44–57.
95. ZhengQ, WangXJ (2008) GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res 36: W358–363.
96. Pérez-LluchS, BlancoE, CarbonellA, RahaD, SnyderM, et al. (2011) Genome-wide chromatin occupancy analysis reveals a role for ASH2 in transcriptional pausing. Nucleic Acids Research 39 : 4628–39 doi: 10.1093/nar/gkq1322
Štítky
Genetika Reprodukční medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 10
-
Všechny články tohoto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



